Approximate time: 45 minutes
Learning Objectives
- Enumerate difficulties with alignment
- Describe the advantages of using the bwa aligner
- Create an
sbatchscript to align reads
The importance of alignment for variant calling
Read alignment is an essential first step in the characterization of DNA sequence variation. This step takes our reads and tries to find the place in the genome where they best fit.
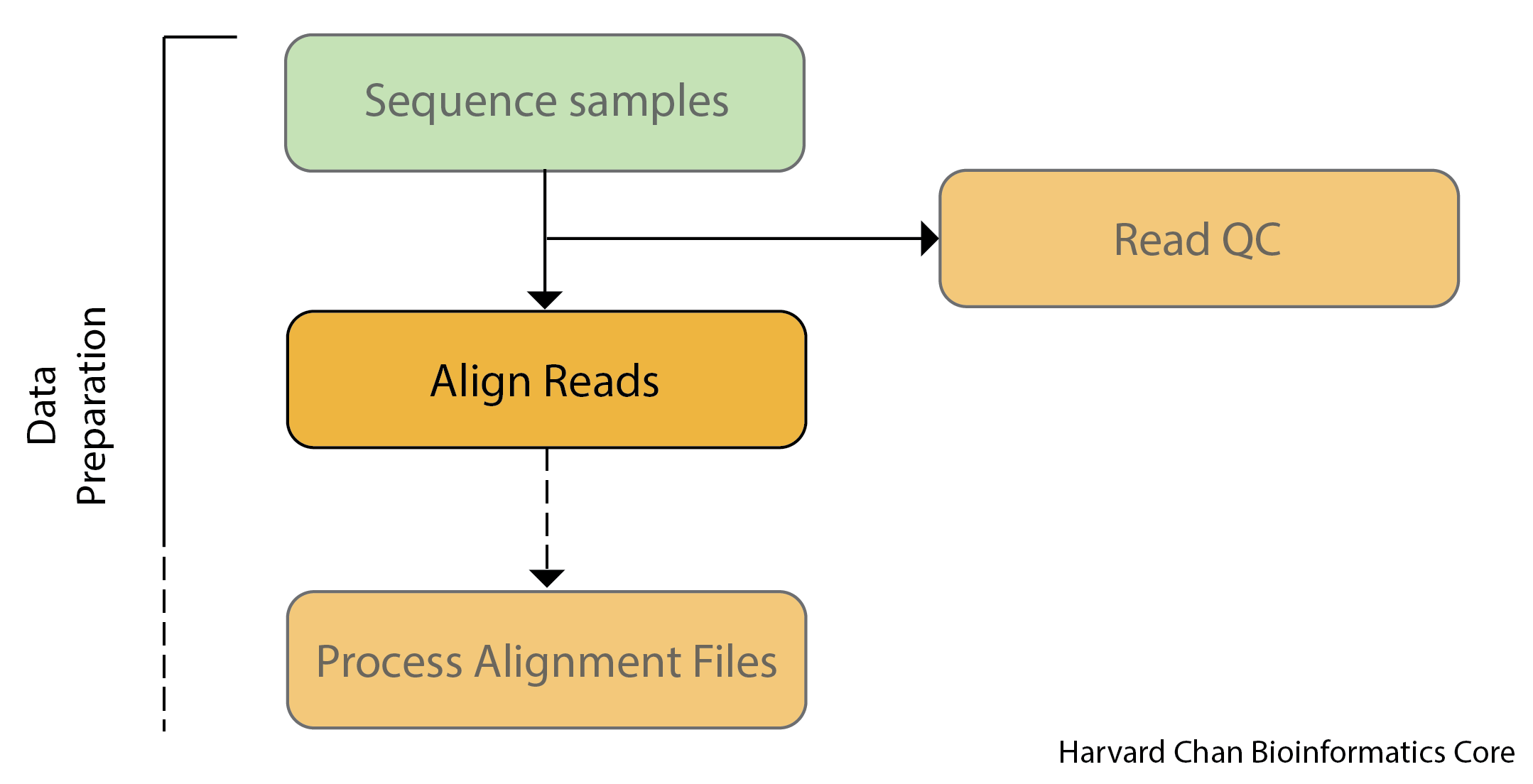
Importantly, the accuracy of variant-calling results can be highly impacted by a low quality of read alignment. In theory, aligning reads sounds like a simple case of string matching to identify where in the genome each read originated from. However, this task is complicated by a multitude of factors including:
- Errors in the sequencing reads
- Errors in the reference genome
- Repetitive areas of the genome
- Variants in the sample
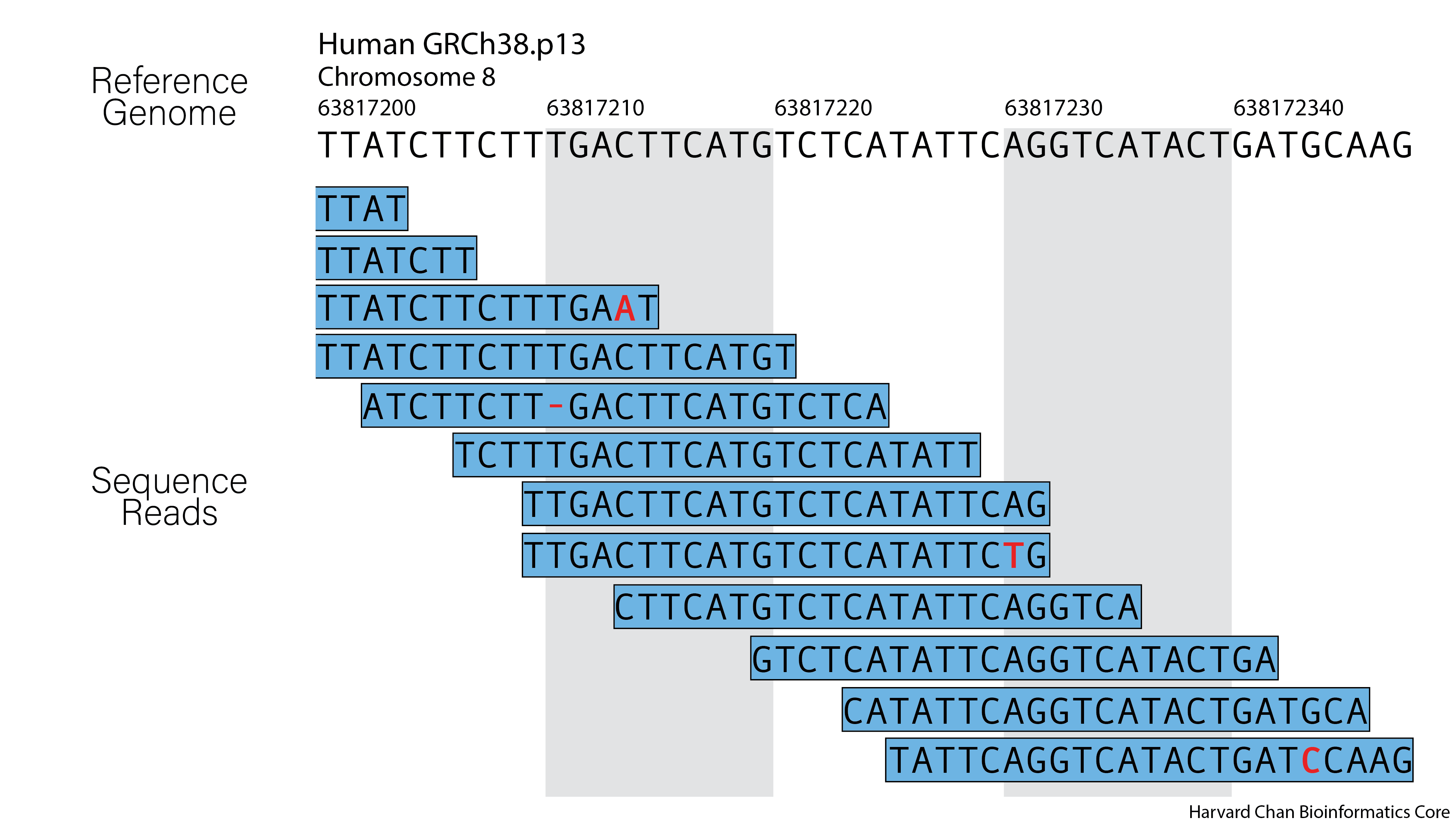
The largest and most difficult part of this task is creating an alignment algorithm that can account for these factors and still provide alignment for millions of reads within a reasonable time frame. There are a multitude of alignment tools availible today; each has strengths and weakness and some are more appropriate for particular types of analysis. In this course, we will be using BWA.
BWA
Many modern alignment tools rely on the Burrows-Wheeler Transform as part of their alignment algorithm; BWA is one of those tools. BWA is a software package for mapping low-divergent sequences against a large reference genome, such as the human genome. It consists of three algorithms: BWA-backtrack, BWA-SW and BWA-MEM. In this workshop we will demonstrate BWA-MEM.
No matter which algorithm you choose, there are two steps to running bwa:
- Create an index of the reference sequence (done once)
- Create a Suffix Array Index
- Generate a Burrows-Wheeler Transform from the suffix array
- Query reads against the index to get alignments
The Burrows-Wheeler Transform and Suffix Arrays are outside of the scope of this course, but a great place to get information on these methods are on Dr. Ben Langmead’s YouTube Channel. Dr. Langmead developed the algorithm for a similar alignment tool, Bowtie, and he has made many useful videos explaining how the components of these alignment tools work.
We have already created an index for you. So we will jump right into running the alignments.
Click here for details on creating your own bwa index
While we will be using an index that has already been made for us, if you need to create an index for a reference sequence using bwa, the steps for this are laid out below.
- Navigate to your reference sequence directory
cd ~/path/to/reference/sequence/directory/
- Create a
bwaindex:bwa index reference_sequence.fasta
This process may take up to 30+ minutes to run depending on the reference sequence size. The output of this command will produce five files:
reference_sequence.fasta.saThis is the binary version of the Suffix Array Indexreference_sequence.fasta.bwtThis is the binary version of the Burrows-Wheeler Transform of the reference sequencereference_sequence.fasta.pacA special binary compression of the reference sequencereference_sequence.fasta.annNotations regarding the reference sequencereference_sequence.fasta.ambNotations regarding base ambiguities (mostly Ns, but also other base ambiguities) in the reference sequence
SAM (Sequence Alignment Map) file format
Alignment information is stored within SAM files. Here, we briefly describe the SAM file, but encourage you to explore this lesson for more in depth detail.
There are two main components to a SAM file are:
- Header section
- This section contains various bits of information pertinent to the file. Some examples include: sequence information (
@SQ), read groups (@RG), history of programs/tools (@PG) used to make the SAM file.
- This section contains various bits of information pertinent to the file. Some examples include: sequence information (
- Alignment section
- Stores alignment data for reads that have attempted to be aligned
- Each row correponds to a single read
- Columns/fields are separated by the tab-delimiter and information on each column/field is briefly described in the image below.
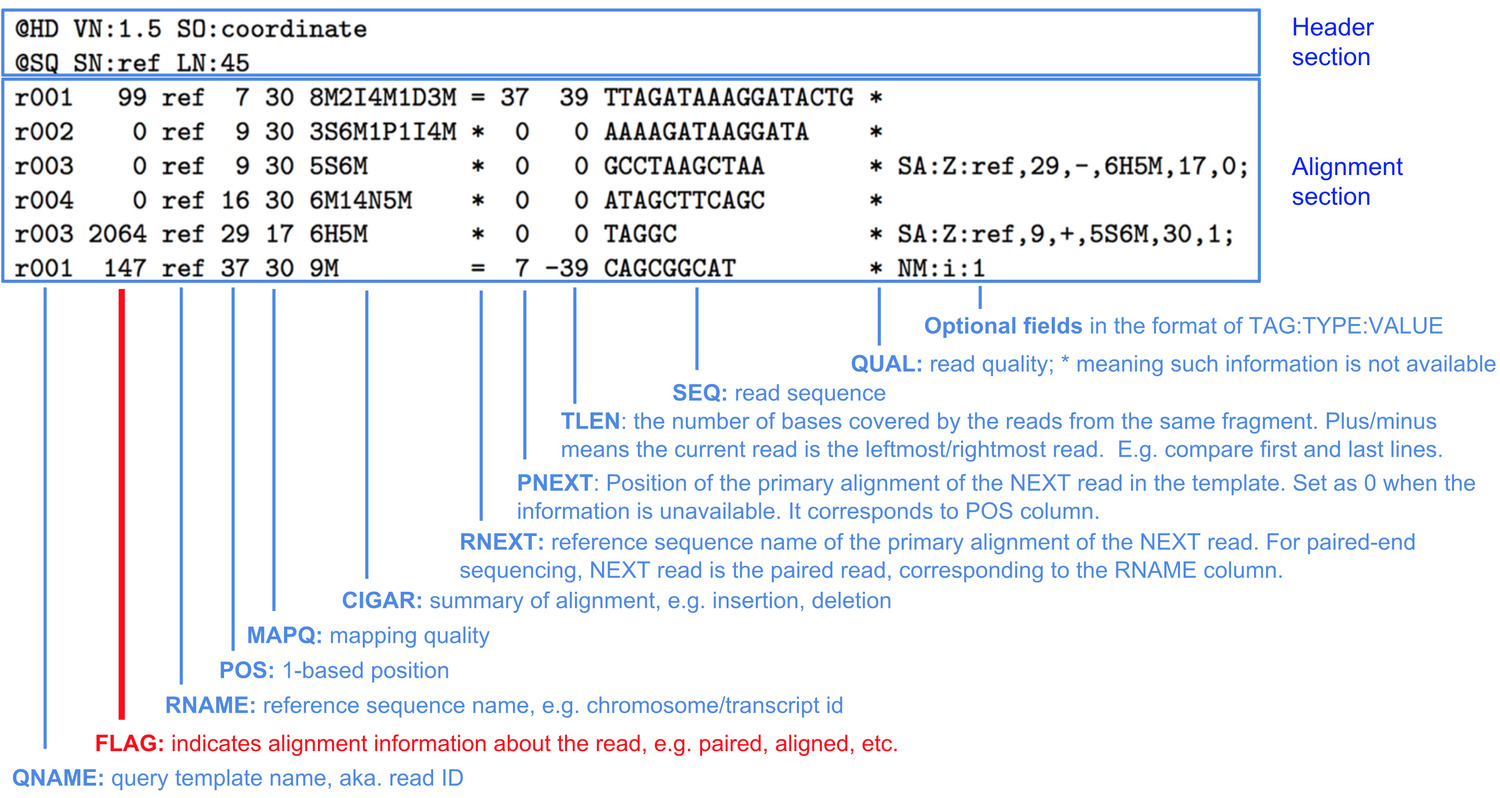
Image source: https://www.samformat.info/sam-format-flag
The bit-wise flags are worth noting here, as they are very helpful for giving the user a rough understanding of the read. Details such as whether the read is paired, has an alignment to the provided reference sequence or is a PCR duplicate can all be encoded into the FLAG. This tool on the Broad’s Website can be very helpful for decoding the SAM FLAGs that you can encounter.
Aligning reads with bwa-mem

BWA-MEM is the latest algorithm of the three bwa algorithms available, and is generally recommended for high-quality queries as it is faster and more accurate. Other features include:
- Performs local alignment (rather than end-over-end)
- Can clip ends of reads, if they do not match
- Can split a read into pieces, mapping each separately (the best aligned piece is then the primary alignment)
- Performs gapped alignment
- Moderate memory requirement (few GB of RAM to hold reference genome)
Let’s begin by looking at the command used to run the alignment, and describing each of the parameters. Do not run the code below, as this just example code.
# DO NOT RUN
# Align reads with bwa-mem
bwa mem \
-M \
-t 8 \
-R "@RG\tID:C6C0TANXX_2\tSM:ZW177\tPL:ILLUMINA\tPU:ZW177" \
/path/to/reference.fa \
LEFT_read_1.fq \
RIGHT_read_2.fq \
-o /path/to/output_file.sam
Let’s breakdown this bwa command.
memThis is the specific algorithm we want to use withinbwa-MThis will mark shorter split hits as secondary-t 8We are going to take advantage of multithreading. In order to do this, we need to specifiy the number of threads that we are going to use as 8.-RThis adds what is called read group information. Some software packages, such asGATK, require read groups, while others are agnostic towards them. However, they can provide important metadata about your reads and thus it is considered best practice to include read group information at the alignment step. This read group information consists of several fields separated by tab-characters (\t), which will be described a little bit later in this lesson.- Reference sequence (indexed)
- Input FASTQ files
-oOutput file name and path (if different from working directory)
A note on paired-end reads: In this case you can see that the reads are paired-end reads because we have provided two FASTQ files as input and they have
_1/_2right before the.fq/.fastqextension.This is a very common annotation for paired-end reads. Alternatively, sometimes they might also be annotated as
_R1/_R2, but once again, they will almost always be placed right before the.fq/.fastqextension.
Read groups
The term Read Group refers to a set of reads that were generated from a single run of a sequencing instrument.
- Simple case: A single library preparation derived from a single biological sample was run on a single lane of a flowcell. All the reads from that lane run belong to the same read group.
- Complex case: When samples are barcoded and pooled, and run across multiple lanes in a flowcell. In this case, each subset of reads originating from a separate library run on that lane will constitute a separate read group.
Read groups are identified in the SAM/BAM file by a number of tags that are defined in the official SAM specification. These tags, when assigned appropriately, allow us to differentiate not only samples, but also various technical features that are associated with artifacts. Having this information allows to take steps towards mitigating the effects of artifacts in downstream analyses [1].
Let’s use the read group from our example bwa command above to demonstrate the use of tags:

- ID: This is the identification for a given batch of reads. This MUST be unique to your experiment.
- SM: This is to mark which sample your reads are coming from. Note, this does not need to be unique like the ID field, since you may have multiple Read Group IDs coming from a single sample.
- PL: This is the platform that the sequencing was run on. For aligning Illumina reads, you should use “illumina” here.
- PU: This is the platform unit and it is ideally supposed to hold
<FLOWCELL_BARCODE>.<LANE>.<SAMPLE_BARCODE><FLOWCELL_BARCODE>is the barcode of the flowcell<LANE>is the lane the data was run on and-
<SAMPLE_BARCODE>is supposed to be a library/sample specific identifer. In some software packages PU can take precedence over the ID field.** If you don’t happen to have the
<FLOWCELL_BARCODE>.<LANE>.<SAMPLE_BARCODE>, just make this field something useful that will help identify the sample. In this case, we didn’t have that information so we will re-use the SM field here.
More information about read groups and some fields we didn’t discuss can be found here.
Exercises
1. The read group field LB (Library) is a required field to when adding read groups using Picard’s AddOrReplaceReadGroups, but we don’t currently have this field in our read group information. How would we alter our bwa command to include LB as well?
2. If we wanted to increase the number of threads used by bwa for processing our alignment to 12, how would we need to modify our bwa command to accommodate this?
Creating a script for alignment
To align reads to the reference sequence, we will need to give ourselves an appropriate amount of resources and so we will need to create an sbatch job submission script. Move into the scripts directory and open up vim.
$ cd ~/variant_calling/scripts/
$ vim bwa_alignment_normal.sbatch
Now, we can copy and paste the following shebang line and sbatch directives into our script file:
#!/bin/bash
# This script is for aligning sequencing reads against a reference genome using bwa
# Assign sbatch directives
#SBATCH -p priority
#SBATCH -t 0-04:00:00
#SBATCH -c 8
#SBATCH --mem 16G
#SBATCH -o bwa_alignment_normal_%j.out
#SBATCH -e bwa_alignment_normal_%j.err
Next, we need to add the modules that we will be using for alignment:
# Load modules
module load gcc/6.2.0
module load bwa/0.7.17
NOTE: On O2, many of the common tools were compiled using
GCCversion 6.2.0, so to be able to access them, we first need to load theGCCmodule.
Finally, we need the bwa command we are going to run. Displayed below is the code we would use specifically for the syn3_normal_1. DO NOT paste this code in your script!
## DO NOT PASTE THIS CODE: EXAMPLE ONLY
bwa mem \
-M \
-t 8 \
-R "@RG\tID:syn3_normal\tPL:illumina\tPU:$SAMPLE\tSM:syn3_normal" \
/n/groups/hbctraining/variant_calling/reference/GRCh38.p7.fa \
~/variant_calling/raw_data/syn3_normal_1.fq.gz \
~/variant_calling/raw_data/syn3_normal_2.fq.gz \
-o /n/scratch/users/${USER:0:1}/${USER}/variant_calling/alignments/syn3_normal_GRCh38.p7.sam
Instead we will introduce bash variables in our script which will allow us to quickly adapt this for use on all other samples we have in our dataset. The variables are described in more detail below:
REFERENCE_SEQUENCE: The path to the bwa indexLEFT_READS: The path to the left read FASTQ file (R1 or 1)RIGHT_READS: The path to the right read FASTQ file (R2 or 2)SAMPLE: The sample prefix (for naming the output file and providing read group information)SAM_FILE: The path and filename for the output alignment file
Another advantage of using bash variables in this way is that it can reduce typos. It can also help make your code more reproducible since you will not be altering the underlying bwa command, which you could inadvertantly do while trying to swap out parameters within your code between scripts if you didn’t use bash variables. Lastly, it can also help make your code more readable to yourself and others.
# Assign files to bash variables
REFERENCE_SEQUENCE=/n/groups/hbctraining/variant_calling/reference/GRCh38.p7.fa
LEFT_READS=/home/$USER/variant_calling/raw_data/syn3_normal_1.fq.gz
RIGHT_READS=`echo ${LEFT_READS%1.fq.gz}2.fq.gz`
SAMPLE=`basename $LEFT_READS _1.fq.gz`
SAM_FILE=/n/scratch/users/${USER:0:1}/${USER}/variant_calling/alignments/${SAMPLE}_GRCh38.p7.sam
NOTE:
$RIGHT_READSuses somebashstring manipulation in order to swap the last parts of their filename. We also usebasenameto parse out the path from a file and when coupled with an argument after the filename, it will trim the end of the filename as well as we can see with the$SAMPLEvariable.
We can now add our command for running bwa and utilize the variables we created above:
# Align reads with bwa
bwa mem \
-M \
-t 8 \
-R "@RG\tID:$SAMPLE\tPL:illumina\tPU:$SAMPLE\tSM:$SAMPLE" \
$REFERENCE_SEQUENCE \
$LEFT_READS \
$RIGHT_READS \
-o $SAM_FILE
Click here to see what our final sbatchcode script for the normal sample alignment should look like
#!/bin/bash # This script is for aligning sequencing reads against a reference genome using bwa
# Assign sbatch directives #SBATCH -p priority #SBATCH -t 0-04:00:00 #SBATCH -c 8 #SBATCH --mem 16G #SBATCH -o bwa_alignment_normal_%j.out #SBATCH -e bwa_alignment_normal_%j.err
# Load modules module load gcc/6.2.0 module load bwa/0.7.17
# Assign files to bash variables REFERENCE_SEQUENCE=/n/groups/hbctraining/variant_calling/reference/GRCh38.p7.fa LEFT_READS=/home/$USER/variant_calling/raw_data/syn3_normal_1.fq.gz RIGHT_READS=`echo ${LEFT_READS%1.fq.gz}2.fq.gz` SAMPLE=`basename $LEFT_READS _1.fq.gz` SAM_FILE=/n/scratch/users/${USER:0:1}/${USER}/variant_calling/alignments/${SAMPLE}_GRCh38.p7.sam
# Align reads with bwa bwa mem \ -M \ -t 8 \ -R "@RG\tID:$SAMPLE\tPL:illumina\tPU:$SAMPLE\tSM:$SAMPLE" \ $REFERENCE_SEQUENCE \ $LEFT_READS \ $RIGHT_READS \ -o $SAM_FILE
Submitting sbatch bwa script
Now your sbatch script for bwa is ready to submit:
$ sbatch bwa_alignment_normal.sbatch
Creating Tumor sbatch script
Because our files will stay mostly the same and we are just swapping the word “normal” for “tumor”, this is an excellent place where we can implement the sed command. The sed command can be used to carry out a variety of tasks, but one of the more popular uses is to use it as a “search-and-replace” tool. A more thorough discussion of sed can be found here. We are going to replace all of the instances of “normal” with “tumor” using a sed command, and redirect the output to a file called bwa_alignment_tumor.sbatch:
$ sed 's/normal/tumor/g' bwa_alignment_normal.sbatch > bwa_alignment_tumor.sbatch
Click here to see what our final sbatchcode script for the tumor sample alignment should look like
#!/bin/bash # This script is for aligning sequencing reads against a reference genome using bwa
# Assign sbatch directives #SBATCH -p priority #SBATCH -t 0-04:00:00 #SBATCH -c 8 #SBATCH --mem 16G #SBATCH -o bwa_alignment_tumor_%j.out #SBATCH -e bwa_alignment_tumor_%j.err
# Load modules module load gcc/6.2.0 module load bwa/0.7.17
# Assign files to bash variables REFERENCE_SEQUENCE=/n/groups/hbctraining/variant_calling/reference/GRCh38.p7.fa LEFT_READS=/home/$USER/variant_calling/raw_data/syn3_tumor_1.fq.gz RIGHT_READS=`echo ${LEFT_READS%1.fq.gz}2.fq.gz` SAMPLE=`basename $LEFT_READS _1.fq.gz` SAM_FILE=/n/scratch/users/${USER:0:1}/${USER}/variant_calling/alignments/${SAMPLE}_GRCh38.p7.sam
# Align reads with bwa bwa mem \ -M \ -t 8 \ -R "@RG\tID:$SAMPLE\tPL:illumina\tPU:$SAMPLE\tSM:$SAMPLE" \ $REFERENCE_SEQUENCE \ $LEFT_READS \ $RIGHT_READS \ -o $SAM_FILE
Once we have created this script we can go ahead and submit it for processing:
sbatch bwa_alignment_tumor.sbatch
NOTE: These scripts may take awhile to run. Each script can take anywhere from 30-45 min to run to completion.
This lesson has been developed by members of the teaching team at the Harvard Chan Bioinformatics Core (HBC). These are open access materials distributed under the terms of the Creative Commons Attribution license (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.