Welcome to the HBC Training Program
We are delighted to have you here!
Are you feeling lost and unsure about bioinformatic anaysis?
- Do you want to utilize high-throughput sequencing data in your research, but not really sure where to start?
- Does the idea of writing your own code for data analysis seem necessary, yet daunting?
- Do you need to brush up on what you already know about analysis of high-throughput sequencing data?
The training team at the Harvard Chan Bioinformatics Core provides bioinformatics training in multiple formats, they can be broadly divided into the following:
- Introduction to High-throughput sequencing (HTS) data analysis series
- Current topics in bioinformatics series
Our current workshops and courses are designed to help biologists become comfortable with using tools to analyse high-throughput data. We are slowly beginning to expand this repertoire to include training for researchers with more advanced bioinformatics skills.
See our current workshop schedule on our training website.
Click on the following questions to expand them for the answers:
What the heck is 'omics?
Over the last 10-15 years many technological advances allow us to assess the entirety of a certain type of molecule(s) in an organsim. The resulting high-throughput data are called 'omics data. We can break 'omics down into 4 specific categories:
- genOMICS - The study of the complete set of DNA in an organism, single cells, or group of cells.
- transcriptOMICS - The study of the complete set of RNA in an organism, single cells, or group of cells.
- proteOMICS - The study of the complete set of Proteins in an organism, single cells, or group of cells.
- metabolOMICS - The study of the complete set of Metabolites in an organism, single cells, or group of cells.
High-throughput data from even a single sample is considered 'omics data. However, we usually are looking at data from large number of biological samples (individuals, cell lines, etc).
What is High-throughput Sequencing (HTS) or Next-generation Sequencing (NGS) data?
- What is a Genome? All of the DNA in an individual or a species
- What is a Transcriptome? All of the RNA in an individual or a species (typically transcribed from DNA in individual cells)
Over the last few years the community is slowly replacing the term NGS (Next-generation Sequencing) with the more descriptive HTS (High-throughput Sequencing).There are hundreds of assays that have been developed for HTS that have enabled us to gain deep insights into the working of a cell. The most commonly used HTS applications that you will encounter are:
- Bulk RNA-seq
- Single-cell RNA-seq
- ChIP-seq
- Whole genome sequencing
- Exome sequencing
- ATAC-seq
- Single-cell ATAC-seq
How do clusters and HPC relate to analysis of HTS data?
Let's return to our book example. If one book is 375,000 characters then 3.2 billion characters (the size of the human genome) translates to 8,533 books! While we might keep tens or even hundreds of books at our house, most people will never have thousands.

Can you imagine dusting this?
It's the same with our local computer. While we might keep small data files on our laptop, we don't want to clutter it up with huge data files. And this is just thinking about storage! Books or data sets need to be organized and kept track of as well. You might be able to alphabetize or organize a hundred books on your own but working with >8,000 books would be overwhelming! The same goes for our computer. To organize billions of base pairs and make sense of our sequencing data we simply need more power. The Mac laptop I am writing this on has 10 cores (a single unit of processing available in our CPU; see below for more information). In comparison, a high perfomance computing (HPC) cluster might have hundreds or thousands of cores. That is a lot more processing capacity, more in line with the large amount of computational work we want to do! Let's take a quick look at the basic architecture of a cluster environment and some cluster-specific jargon.
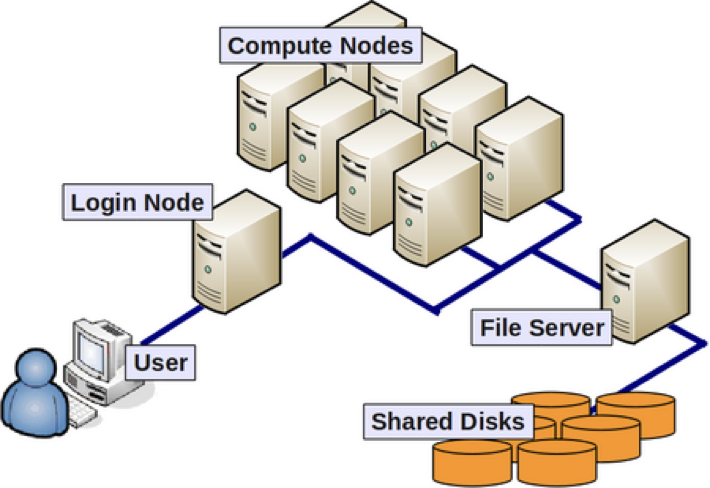
- More memory (temporary storage)
- Many more, faster CPUs
- Each of those CPUs has many more cores
Why use the cluster or an HPC environment?
- A lot of software is designed to work with the resources on an HPC environment and is either unavailable for, or unusable on, a personal computer.
- If you are performing analysis on large data files (e.g. high-throughput sequencing data), you should work on the cluster to avoid issues with memory and to get the analysis done a lot faster with the superior processing capacity. Essentially, a cluster has:
- 100s of cores for processing!
- 100s of Gigabytes or Petabytes of storage!
- 100s of Gigabytes of memory!
Parallelization
Point #2 in the last section brings us to the idea of parallelization or parallel computing that enables us to efficiently use the resources available on the cluster.One input file
Let's start with the most basic idea of processing 1 input file to generate 1 output (result) file. On a personal computer this would happen with a single core in the CPU.

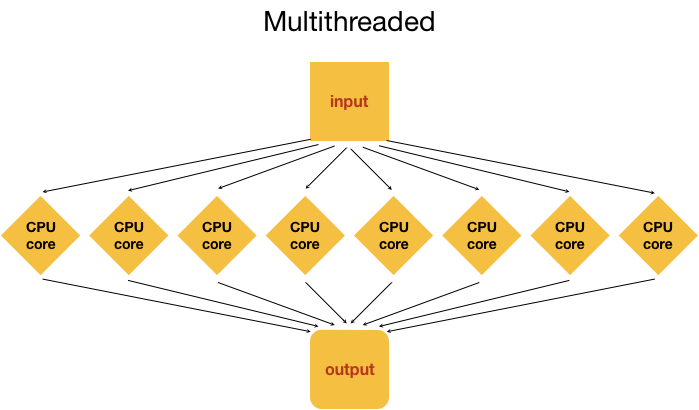
Note: Multithreading is done internally by analysis tools being employed, and not by manually splitting the input (except in very unusual circumstances).
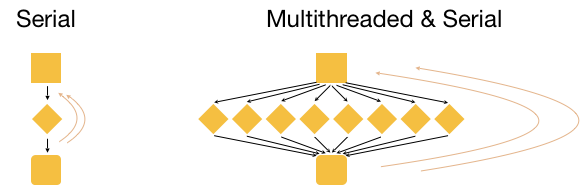
Three input files
Now, what if we had 3 input files? Well, we could process these files in serial, i.e. use the same core(s) over and over again, as shown in the image below.
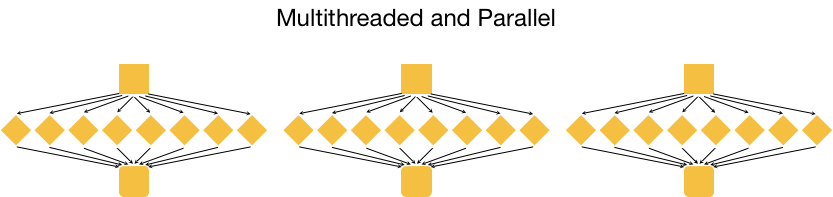
What is shell and how does it relate to clusters?
So how might you actually use a cluster? Unfortunately you can't just walk up to where the cluster is stored and start using it. Clusters are accessed remotely, that means that you connect to the cluster from your own computer. You will do this from the command line or a text-based user interface. We are used to clicking on applications we want to use and selecting various commands from dropdown menus. Clusters do not work this way. Any task that you want a cluster to do has to be communicated through a text command.

The FAS-RC Cluster
If you have never taken a computer science course or worked with clusters before this will all be brand new to you. But don't worry, we have courses for that! For now let's just review the basics. To look at command line on your own computer you can open the Terminal program on Macs or for Windows download Git BASH or similar application. The shell is what runs in these programs to interpret your commands. These programs all use Bash, a command language. As you get into HTS and computational work you will encounter a lot of languages such as Python, Perl, Fortran, R, C++, Java and more. You can think of these as being akin to human languages; French and English sound very different and have different syntax (the order of words) but can be used to convey the same message. At HBC training we recommend that you become familiar (or fluent) in bash and R to begin with.What is R and what can it do?
Why do we recommend R instead of other languages? According to R-project, "R is a language and environment for statistical computing and graphics." R is also a well developed and relatively simple language that is widely used among data scientists and people in STEM. Compelling arguements for learning R include:
- It’s open-source. This means no fees or licenses are needed and you won't get any pop ups asking for money.
- It’s platform-independent. This means that R runs on all operating systems (Mac, Windows, Linux) and R scripts written on on platform can be run on any other platform.
- People write packages for R, especially in the field of bioinformatics. The R language has more than 10,000 packages stored in the CRAN repository, and that number is continuously increasing. Many packages for analyzing HTS data are written for R such as DESeq2 and Seurat among others.
- Data wrangling, i.e., turning raw data into the desired format. Data wrangling is necessary for working with any 'omics data set and R has many packages that can turn unstructured, messy data into a structured format.
- Great plotting programs. R has wonderful packages to make publication ready figures. We even have a workshop devoted to it!
- It’s great for statistics. Unlike SAS which is very costly, R is free and has many different statistical packages available.
- You can use R for Machine Learning. R is ideal for machine learning operations such as regression and classification and even for artificial neural network development.
- R is growing. R has a solid support program and help with issues is widely available. New packages and features are available regularly!
Where do I go from here?
Hopefully you now feel like you have a grasp on some of these terms. If you want to start getting your hands wet, we recommend that you take our Intro to R Course and the appropriate shell intro for the cluster you will use, either O2 or FAS-RC. You are free to take a workshop with us or work through the lessons yourself at your own pace. See our below for all of our offerings.
NOTE: The tables below are also represented using a schematic figure which can be found on this page here.
Introduction to high-throughput sequeuncing (HTS) data analysis series:
This series of workshops is divided into 2 categories, Basic Data Skills and Advanced Topics. The Basic workshops serve as the foundation that participants can build upon in the Advanced workshops and we will be offering these as pairs with the appropriate basic workshop preceding an advanced one. Please see below for a description of workshops under each of these two categories.
Basic Data Skills:
These workshops provide an introduction to computational skills required for someone to get started with analyzing high-throughput sequencing data independently. These have no prerequisites and do not require any prior experience with programming.
| Topic and Link(s) to lessons | Prerequisites |
|---|---|
| Shell for Bioinformatics - O2 cluster | None |
| Introduction to R | None |
| Introduction to R (video tutorials) | None |
Advanced Topics:
These are intensive workshops that instruct participants on how to design experiments, and efficiently manage & analyze data. They focus on the workflow for a specific type of next-generation sequencing application (i.e RNA-seq, ChIP-seq). These workshops require participants to have taken one or more of the Basic Data Skills workshops as listed in the table below.
| Topic and Link(s) to lessons | Prerequisites |
|---|---|
| Introduction to bulk RNA-seq: From reads to count matrix - O2 cluster | Shell for Bioinformatics |
| Introduction to Differential Gene Expression Analysis | Introduction to R |
| Investigating chromatin biology using ChIP-seq and CUT&RUN - O2 cluster | Shell for Bioinformatics |
| Introduction to single cell RNA-seq | Introduction to R |
| Introduction to Variant Analysis | Shell for Bioinformatics |
| Tools for Reproducible Research | Introduction to R |
| Pseudobulk and related approaches for scRNA-seq analysis | Introduction to R |
| Introduction to Peak Analysis | Introduction to R |
Current topics in bioinformatics series:
These workshops provide instruction on basic data skills as well as introduce new topics of interest to the community.
R-based short workshops:
| Topic and Link(s) to lessons | Prerequisites |
|---|---|
| Foudations in R | None |
| Practical Applications of R | Beginner R or Completion of the Intro to R online resource |
| Functional analysis of gene lists | Beginner R or Intro to R workshop |
| Reproducible Research using RMarkdown | Beginner R or Intro to R workshop |
| Publication Perfect I: Data visualization basics with ggplot2 | Beginner R or Completion of the Intro to R online resource |
| Publication Perfect II: Figure formatting in R | Publication Perfect: Part I |
| Interact with your data using RShiny | Beginner R or Completion of the Intro to R online resource |
Shell-based short workshops:
| Topic and Link(s) to lessons | Prerequisites |
|---|---|
| Foundations in Shell | None |
| Intermediate Shell/Accelerate with Automation | Basic Shell |
| Advanced Shell/Finding and Summarizing Data from Colossal Files | Basic Shell |
| Tips and Tricks on O2 | Basic Shell |
| “Track Changes” for Your Code: An Introduction to Git and GitHub | No pre-requisite (GitHub Desktop) |
| Coding with Others: Managing Conflicts on GitHub | “Track Changes” for Your Code |
| Accessing genomic reference and experimental sequencing data | Basic Shell |
Other short workshops:
| Topic and Link(s) to lessons | Prerequisites |
|---|---|
| Introduction to Python | None |
| Planning a bulk RNA-seq analysis: Part I | None |
| Planning a bulk RNA-seq analysis: Part II | None |
| Make your (RNA-seq) data analysis reproducible- Taught by Julie Goldman from Countway Library | None |
| Improving your (RNA-seq) data analysis using version control (Git) - In collaboration with HBC-RCS | None |
Contact us:
Email: hbctraining@hsph.harvard.edu
Webpage: http://bioinformatics.sph.harvard.edu/training/
These materials have been developed by members of the teaching team at the Harvard Chan Bioinformatics Core (HBC) RRID:SCR_025373. These are open access materials distributed under the terms of the Creative Commons Attribution license (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
A lot of time and effort went into the preparation of these materials. Citations help us understand the needs of the community, gain recognition for our work, and attract further funding to support our teaching activities. Thank you for citing the corresponding course (as suggested in its “Read Me” section) if it helped you in your data analysis.