Welcome to the HBC Training Program
We are delighted to have you here!
The training team at the Harvard Chan Bioinformatics Core provides training to help biologists become comfortable with using bioinformatics tools to analyse high-throughput sequencing (HTS) data.
We offer courses and skills at three different levels starting at the basics and building upwards. We focus on the two most commonly used HTS interfaces, R and Bash/Shell.
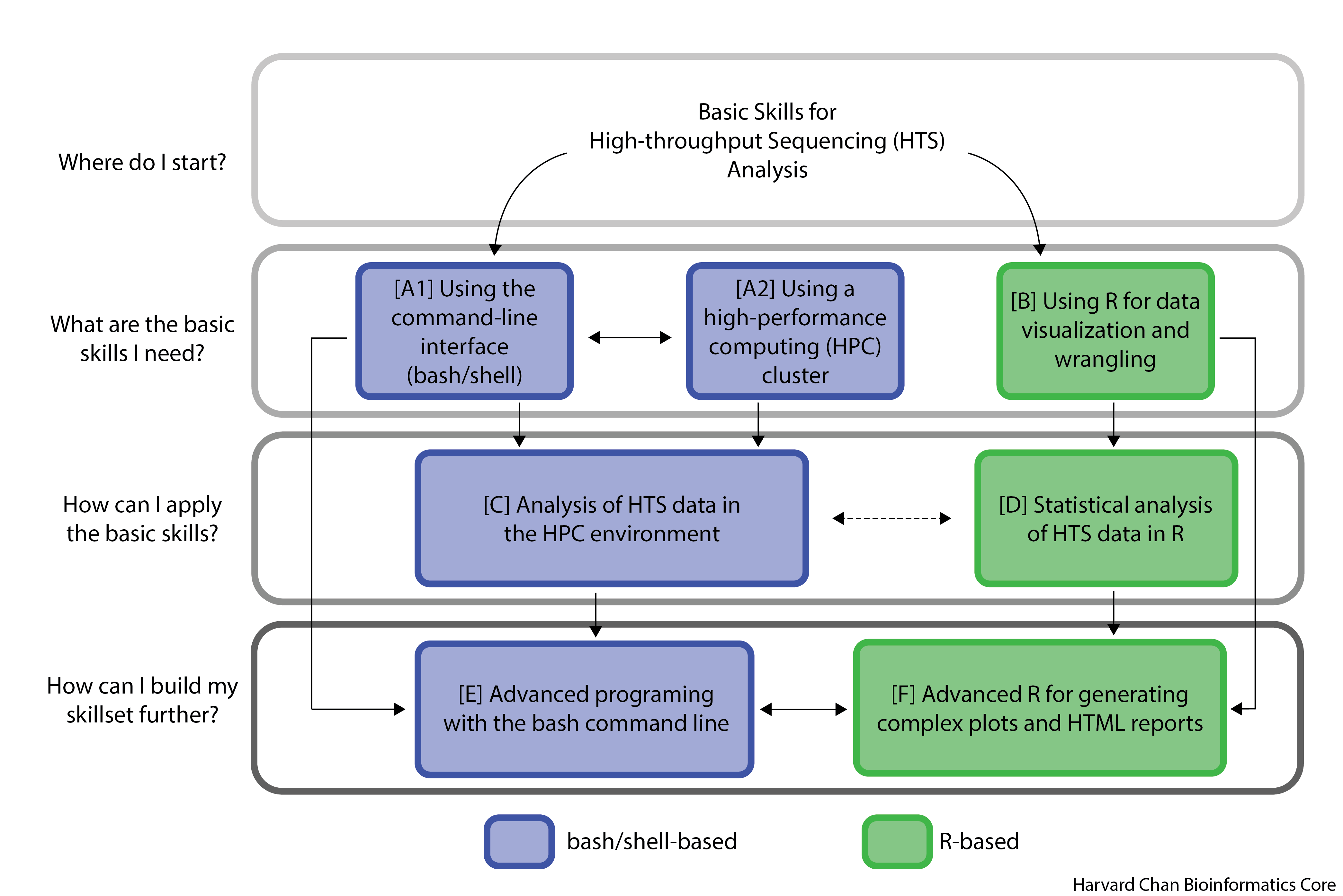
What are the basic skills I need?
| Skill | Who needs it | Overview | Courses |
|---|---|---|---|
| A1 - Using the command line interface | Anyone planning on doing scientific computing using the command-line. | Understanding the need for shell and master basic commands | |
| A2 - Using a HPC cluster | Anyone who wants to efficiently run analyses on large datasets (requiring more computational resources than a laptop can provide). | Understanding the components of a high performance compute cluster (HPC), and learning to navigate and properly use available HPCs at Harvard. | |
| B - Using R | Anyone who wants to learn a programming language that is especially useful for data wrangling and statistics. | Learning the R and RStudio interface, Basic R syntax, and data visualization |
How can I apply the basic skills?
| Skill | Who needs it | Overview | Courses |
|---|---|---|---|
| C - Analysis of HTS data in the HPC environment | Anyone planning on doing genomic or transcriptomic next-generation sequencing and is interested in analyzing their own data. | - Analysis of bulk RNAseq data, Variant Analysis, and sequencing data related to Chromatin biology starting with raw data. - Automating the workflow with advanced shell scripts. |
|
| D - Statistical Analysis of HTS data in R | Anyone who wants to use popular R packages for downstream analysis of HTS data. Main focuses include Seurat and DESeq2. | - Using R to implement best practices workflows for the analysis of various forms of HTS data. - Clear explanations of the theory behind each step in of the workflow. |
How can I build my skillset further?
| Skill | Who needs it | Overview | Courses |
|---|---|---|---|
E - Advanced programming with the bash command line |
Anyone who wants to create custom shell scripts and utilize bash for various tasks. | Learning to include version control in your projects and advanced bash scripting | |
| F - Advanced R for generating complex plots and reports | Anyone who wants to make publication quality figures Anyone who wants to make high level HTML reports of analyses |
Exploring additional R features such as reports and publication perfect figures |
Additional Courses
Contact us:
Email: hbctraining@hsph.harvard.edu
Webpage: http://bioinformatics.sph.harvard.edu/training/
These materials have been developed by members of the teaching team at the Harvard Chan Bioinformatics Core (HBC) RRID:SCR_025373. These are open access materials distributed under the terms of the Creative Commons Attribution license (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
A lot of time and effort went into the preparation of these materials. Citations help us understand the needs of the community, gain recognition for our work, and attract further funding to support our teaching activities. Thank you for citing the corresponding course (as suggested in its “Read Me” section) if it helped you in your data analysis.