Learning Objectives
- Discuss the advantages of utilizing job dependencies
- Implement a job dependency
- Cancel a job that is running on the cluster
- Monitor submitted jobs
Overview
- Job dependencies
- Job arrays
- Checking on your jobs in queue with
squeueandscontrol - Getting additional information about your jobs with
sacctandO2_jobs_report - Canceling your job(s) with
scancel - Keeping track of time
- Running jobs in the background with
&,bg/fg, andscreen - O2 Portal
Managing jobs on the cluster
Most, if not all, high performance compute clusters (HPCs) utilize a job scheduler. As we have previously discussed, O2 uses one of the most popular ones, SLURM. These job schedulers aim to allow for fair use of limited computational resources in a shared network. The purpose of this lesson is to introduce you to various ways of effectively setting up and managing your jobs on an HPC environment.
Job dependencies on SLURM
Before we get to more basic commands on how to monitor our running jobs, we will introduce a few concepts that are helpful for scenarios that go beyond the simple job submission script.
Let’s begin with the idea that perhaps one of your jobs is dependent on the completion of another job. Do you sit there for 48 hours (or however long it takes to run to completion) before you set up the next job to run? Do you add all the commands for you workflow into a single script to run serially, even though each step requires different resources?
The answer to each of the questions above, is no.
The solution is to have your jobs queue in such a way that once one job finishes, it automatically queues the next job.
Job dependencies are very useful when you:
- Have consecutive jobs you want to run
- Don’t want to/have the time to manage the submission of consecutive jobs
NOTE: Job dependencies are not unique to SLURM, many other job schedulers, like PBS, also have a feature similar to this!
The syntax for using job dependencies in SLURM is implemented as an SBATCH directive/option called --dependency.
This option has several arguments that it can accept, but the most commonly used one is afterok.
sbatch --dependency=afterok:<job_ID_number> job_2.sbatch
Using afterok means that after the following job ends without an error, then it will remove the hold on the dependent job. After the afterok part you can separate one or more jobs with colons to signify which jobs are dependent.
Multiple jobs can be dependent on a single job. Conversely, we can have a single job dependent on multiple jobs. Both of these examples (and their associated commands) are shown in the figure below.
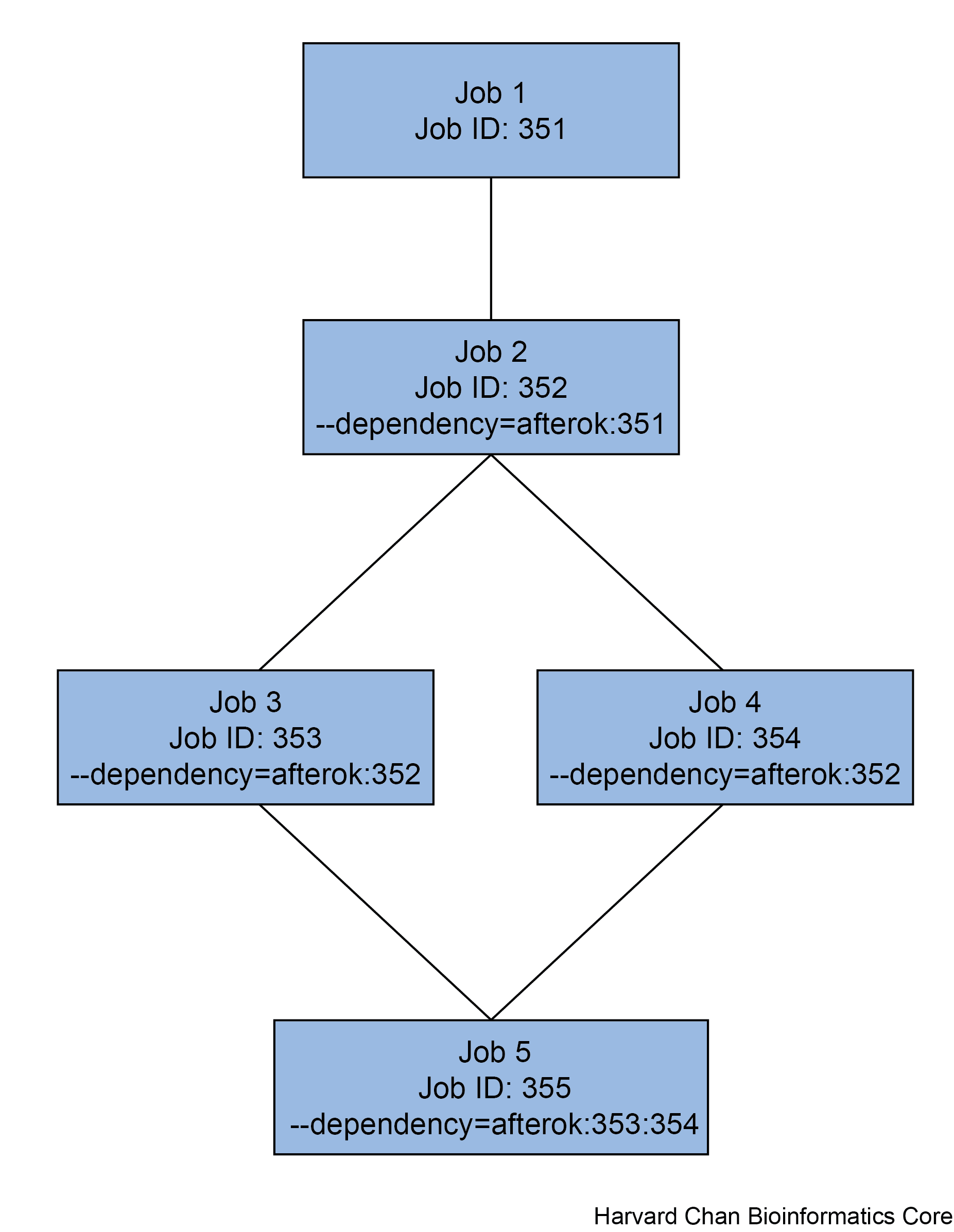
Example of job dependencies
Let’s go to our scratch space to test this out!
cd_scratch
Move in to the sleep_scripts directory that we downloaded earlier:
cd sleep_scripts
We can inspect these two scripts by using the cat command:
cat job_1.sbatch
cat job_2.sbatch
They are very basic scripts that contain the sleep command. Each script simply pauses for 180 seconds and 30 seconds, in jobs 1 and 2, respectively. Before we run these scripts, use vim to modify both of these scripts to have your e-mail address in the line shown below. In this way, you will receive the email notification on the status of these jobs.
#SBATCH --mail-user=Enter_your_email_here
Next, go ahead and submit job_1.sbatch to the cluster:
sbatch job_1.sbatch
At the command prompt you should observe the following text:
Submitted batch job <job_ID_number>
Use the job_ID_number which was returned from the first script as a dependency when submitting your second script job_2.sbatch. Make sure to use the afterok option as we show below:
sbatch --dependency=afterok:<job_ID_number> job_2.sbatch
Now, the second job will only begin after the completion of the first. If you want to take a quick look at your jobs with squeue:
squeue -u $USER
You will notice that job_1 is hopefully running, while job_2 has a “PENDING” state and the “NODELIST(REASON)” states that it is due to a “(Dependency)”.
NOTE: While the behavior can change between implementations of SLURM, on O2, when a job exits with an error, it removes all
afterokdependent jobs from the queue. Some other implementations of SLURM will not remove these jobs from the queue, but the provided reason when you check will beDependencyNeverSatified. In this case, you will need to manually cancel these jobs.
Job arrays
If your work consists of a large number of tasks which differ only in some parameter, you can conveniently submit many tasks at once using a job array. There are two important components to note:
- The individual tasks in the array are distinguished by an environment variable,
$SLURM_ARRAY_TASK_ID, which Slurm sets to a different value for each task. - You set the range of values with the
--arrayparameter.
Let’s look at a very simple example adapted from the technical documentation wiki of the Digital Research Alliance of Canada:
## DO NOT RUN - EXAMPLE ONLY
#!/bin/bash
#SBATCH --array=1-10
#SBATCH --time=3:00:00
<command 1>
<command 2> ...
This job will be scheduled as ten independent tasks.
- Each task has a separate time limit of 3 hours.
- Each task may start at a different time on a different node.
What is stored in the $SLURM_ARRAY_TASK_ID variable?
This variable takes on the numbers 1 through 10 (from --array=1-10) and is used to assign the individual tasks a job ID. In the commands that follow, we can use the $SLURM_ARRAY_TASK_ID value stored as the distinguishing factor between the 10 tasks.
For example, we can use the values 1-10 to identify 10 different samples that we want to run the software on:
software_program_x mydataset_sample_${SLURM_ARRAY_TASK_ID}
Alternatively, we might want to run the the software on the same sample but with each task changing a specific parameter value:
software_program_y $SLURM_ARRAY_TASK_ID some_argument another_arg
There are also various ways in which we can use the $SLURM_ARRAY_TASK_ID numbers to access a text file which contains the values we need for each task. Using a job array instead of a large number of separate serial jobs has advantages for you and other users! It increases efficiency and you spend less time waiting, but the scheduler does not have to analyze job requirements for each task in the array separately, so it can run more efficiently too.
NOTE: You should not use a job array to submit tasks with very short run times, e.g. much less than an hour. These tasks are better off run using a shell loop inside a job.
You can learn more about implementing Slurm arrays in our Accelerate with automation: Arrays in Slurm lesson.
Monitoring your jobs
As your jobs are running, there are various commands you can use to check in on them and see how things are going. If things are not okay, there is also opportunity for you to intervene if necessary.
Checking jobs in the queue using squeue
Earlier we had used the squeue command to check on our jobs. This command is used to view information about jobs located in the SLURM scheduling queue. We had demonstrated that we could provide the -u $USER argument to print only $USER’s jobs. A shortcut to getting the same output would be:
squeue --me
Alternatively, HMS-RC has created a simplified command which returns the results of squeue --me with some additional columns of information related to resources as shown below:
O2squeue
This returns:
JOBID PARTITION STATE TIME_LIMIT TIME NODELIST(REASON) ELIGIBLE_TIME START_TIME TRES_ALLOC
21801263 interactive RUNNING 12:00:00 2:09:52 compute-a-16-160 2020-11-09T11:35:49 2020-11-09T11:36:19 cpu=1,mem=2G,node=1,billing=1
Using scontrol for detailed information
When you use squeue, sometimes you don’t get all of the information that you might like. The scontrol command can help give you a more detailed picture of the job submission. The syntax for using scontrol is:
scontrol show jobid <job_ID_number>
It will return an output that looks like:
JobId=Job_ID JobName=job_name.sbatch
UserId=$USER(XXXXXXX) GroupId=$USER(XXXXXXX) MCS_label=N/A
Priority=369878 Nice=0 Account=Account_name QOS=normal
JobState=RUNNING Reason=None Dependency=(null)
Requeue=0 Restarts=0 BatchFlag=1 Reboot=0 ExitCode=0:0
RunTime=00:40:53 TimeLimit=1-00:00:00 TimeMin=N/A
SubmitTime=2024-05-06T22:38:07 EligibleTime=2024-05-06T22:38:07
AccrueTime=2024-05-06T22:38:07
StartTime=2024-05-06T22:38:31 EndTime=2024-05-07T22:38:31 Deadline=N/A
SuspendTime=None SecsPreSuspend=0 LastSchedEval=2024-05-06T22:38:31 Scheduler=Backfill
Partition=medium AllocNode:Sid=compute-e-16-230:17259
ReqNodeList=(null) ExcNodeList=(null)
NodeList=compute-a-16-163
BatchHost=compute-a-16-163
NumNodes=1 NumCPUs=1 NumTasks=1 CPUs/Task=1 ReqB:S:C:T=0:0:*:*
ReqTRES=cpu=1,mem=64G,node=1,billing=5
AllocTRES=cpu=1,mem=64G,node=1,billing=5
Socks/Node=* NtasksPerN:B:S:C=0:0:*:* CoreSpec=*
MinCPUsNode=1 MinMemoryNode=64G MinTmpDiskNode=0
Features=(null) DelayBoot=00:00:00
OverSubscribe=OK Contiguous=0 Licenses=(null) Network=(null)
Command=/Path/to/submission/script/job_name.sbatch
WorkDir=/Path/where/script/was/submitted/from
StdErr=/Path/where/script/was/submitted/from/Job_ID.err
StdIn=/dev/null
StdOut=/Path/where/script/was/submitted/from/Job_ID.out
Power=
As you can see, there is a lot of information about the job reported back to you. It tells you the job’s state, when it started, when the job will end if it doesn’t finish early, the compute node that it is on, partition used, any job dependencies it has, the resources requested, where the standard error and standard output is written to. Almost any question you might have about a job can be answered within here. You can also use scontrol to check information about a job that has already completed.
Checking on all submitted jobs using sacct or O2_jobs_report
The sacct command displays information on jobs, job steps, status, and exitcodes by default (these are jobs completed in addition to those in the queue):
sacct
Your output might look something like:
JobID JobName Partition Account AllocCPUS State ExitCode
------------ ---------- ---------- ---------- ---------- ---------- --------
38003155 bash interacti+ bcbio 1 CANCELLED+ 0:0
38003155.ex+ extern bcbio 1 COMPLETED 0:0
38003155.0 bash bcbio 1 CANCELLED 0:9
38003191 job_1 short bcbio 1 CANCELLED+ 0:0
38003191.ba+ batch bcbio 1 CANCELLED 0:15
38003191.ex+ extern bcbio 1 COMPLETED 0:0
38003285 job_1 short bcbio 1 CANCELLED+ 0:0
38003285.ba+ batch bcbio 1 CANCELLED 0:15
38003285.ex+ extern bcbio 1 COMPLETED 0:0
38003385 bash interacti+ bcbio 1 CANCELLED+ 0:0
38003385.ex+ extern bcbio 1 COMPLETED 0:0
38003385.0 bash bcbio 1 CANCELLED 0:15
38003495 bash interacti+ bcbio 1 RUNNING 0:0
38003495.ex+ extern bcbio 1 RUNNING 0:0
38003495.0 bash bcbio 1 RUNNING 0:0
This can give you a nice overview of your recently completed jobs. It gives you information about the jobs that are running or pending. However, there is a jobs report that was developed by the folks at HMS-RC that is more informative and the command is called O2_jobs_report. Let’s take a look at it:
O2_jobs_report
The output will look something like:
JOBID USER ACCOUNT PARTITION STATE STARTTIME WALLTIME(hr) RUNTIME(hr) nCPU,RAM(GB),nGPU PENDINGTIME(hr) CPU_EFF(%) RAM_EFF(%) WALLTIME_EFF(%)
-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
37960930 <user> bcbio interactive COMPLETED 2024-05-09 9.0 1.6 1,1.0,0 0.01 0.1 1.2 17.9
37962316 <user> bcbio priority COMPLETED 2024-05-09 2.0 0.0 1,8.0,0 0.0 93.8 49.6 1.3
37976042 <user> bcbio interactive FAILED 2024-05-09 12.0 1.9 1,4.0,0 0.01 0.0 0.1 15.5
38003155 <user> bcbio interactive CANCELLED 2024-05-10 12.0 0.0 1,4.0,0 0.01 2.2 0.1 0.3
38003191 <user> bcbio short CANCELLED 2024-05-10 0.1 0.0 1,0.0,0 0.02 0.0 0.0 14.7
38003285 <user> bcbio short CANCELLED 2024-05-10 0.1 0.0 1,0.0,0 0.03 0.0 0.0 12.0
This is an excellent way to not only get the same information that sacct provides, but also it gives the user better context about the CPU, memory and time efficiency of their requested jobs. This can very helpful for users to know how they can more responsibly use requested resources in their future jobs.
Canceling your job(s) with scancel
There might be a time that you want to cancel a job that you’ve started. The command that you can use to cancel a job is scancel and the syntax looks like:
# DO NOT RUN
scancel <job_id>
The job_id is the job_id that you get when you submit a job or you can find it out by looking at your submitted jobs with using O2squeue or squeue --me.
If you have many jobs that you need to be cancelled, there is a useful shortcut for running scancel across all of your jobs:
# DO NOT RUN
scancel --me
This will cancel all of your jobs, including any interactive jobs you might be running. We can test this out by submitting our job_1.sh to the cluster twice:
sbatch job_1.sbatch
sbatch job_1.sbatch
We should be able to see that we now have two job_1.sbatch scripts submitted when we use our O2squeue:
O2squeue
Now let’s cancel them both:
scancel --me
Now we should see that we have no jobs running when we check O2squeue:
O2squeue
Keeping Track of Time
There are times when you want to keep track of what is going on. If you are able to see how long a task takes to run - you can easily plan for your requested resources on a future run.
watch
Sometimes one may want to see the output of a command that continuously changes. The watch command is particularly useful for this. Add watch before your command and your command line will take you to an output page that will continually update your command. Common uses for watch include:
1) Viewing as files as they created:
watch ls -lh <directory>
2) Monitoring jobs on the cluster:
watch squeue --me
The default interval for update is two seconds, but that can be altered with the -n option. Importantly, the options used with watch command need to be placed before the command that you are watching or else the interpreter will evaluate the option as part of the watched command’s options. An example of this is below:
# Update the watch every 4 seconds
watch -n 4 squeue --me
time
Sometimes you are interested in knowing how long a task takes to complete. Similar to the watch command you can place time in front of a command and it will tell you how long the command takes to run. This can be particularly useful if you have downsampled a dataset and you are trying to estimate how long the full dataset will take to run. An example can be found below:
time ls -lh
The output will have three lines:
real 0m0.013s
user 0m0.002s
sys 0m0.007s
realis most likely the time you are interested in since it displays the time it takes to run a given command.userandsysrepresent CPU time used for various aspects of the computing and can be impacted by multithreading.
Running jobs in the background
Sometimes you may start a command that will take a few minutes and you want to have your command prompt back to do other tasks while you wait for the initial command to finish. There are various ways to do this.
-
One way is to add the
&argument to the end of your command when you first type it. -
If you’ve started your job already but forgot to include the
&argument, usebg. To usebg, you will need to know the following:- Pause the command with Ctrl + Z.
- Send the command to the background with the command
bg. When you do this the command will continue from where it was paused. - If you want to bring the task back to the foreground, you can use the command
fg.
In order to test these methods, we will briefly re-introduce the sleep command. sleep just has the command line do nothing for a period of time denoted in seconds by the integer following the sleep command. This is useful if you want a brief pause within a loop, such as between submitting a bunch of jobs to the cluster. The syntax is:
# DO NOT RUN
sleep [integer_for_time_in_seconds]
So if you wanted there to be a five second pause, you could use:
sleep 5
Now that we have re-introduced the sleep command let’s go ahead and run it for 180 seconds to simulate a task that is running that might take a few minutes to run.
If we know in advance we want to do other things while we’re running our command, we can simply add the & argument:
sleep 180 &
You’ll notice you can type other commands freely even though sleep is running in the background. For now, let’s cancel this command by typing Ctrl + X + C.
Now, what if we started sleep and didn’t add & to our command?
sleep 180
Type Ctrl + Z and this will pause that command. Then move that task to be running in the background with:
bg
The sleep command is now running in the background and you have reclaimed your command-line prompt to use while the sleep command runs. You’ll also notice that it added the & to your sleep command in the terminal output.
If you want to bring the sleep command back to the foreground, type:
fg
fgwill also work for commands which used the&.
These commands are useful whenever you are running commands/scripts that take a few minutes to run but don’t have super large processing requirements. Some examples include:
- Indexing a FASTA file
- Executing a long command with many pipes
- Testing a command interactively
Typically, lengthy tasks should be submitted as jobs to the cluster. However, you will find it is not uncommon to be running an interactive session and wanting to concurrently have access to the command prompt to do other things.
screen
An alternative to using & and bg, is to use screen, which allows the user to have multiple command-line interfaces open simultaneously while on the cluster. So instead of running something in the background or on a different terminal window, the user can just place it in a different screen and it is easy to navigate between screens. Let’s start by copying a screens template configuration file that HMS-RC provides over to our home directory:
cp /n/shared_db/misc/rcbio/data/screenrc.template.txt ~/.screenrc
And we can look to see what this template using less:
less ~/.screenrc
And the contents are:
hardstatus alwayslastline
hardstatus string "%{.bW}%-w%{.rW}%n %t%{-}%+w %=%{..G} %H %{..Y} %m/%d %C%a "
# Default screens
screen -t test1 1
screen -t test2 2
screen -t run1 3
screen -t run2 4
#screen -t other 5
# replace Crtl+a by Ctrl + x
#escape ^Xx
Within this configuration file, you can see the number of default windows are 4, but this can be modified with the addition or removal of screen -t lines. Once you have the screen configuration file copied to your home directory, you won’t need to do it again and to use screen, you simply need to type:
screen
Your terminal window should now look like:
In order to navigate between your screens you can type Ctrl + a + insert_screen_number. For example, if we wanted to jump to the second screen, we can type Ctrl + a + 2. Let’s go ahead and get a sleep command running:
sleep 60
And now let’s hop to the first screen while we wait on it to run with Ctrl + a + 1.
Now, you might be thinking, “If I am using Ctrl + a + insert_screen_number to jump between screens, what happens to the Ctrl + a that we use to jump to the beginning of our command-line?” We can use Ctrl + a + a to jump to the beginning of the command-line when inside of screens, however Ctrl + e remains for jumping to the end of the line.
We can exit screen by using Ctrl + d. A few useful keyboard shortcuts for screen are listed in the table below:
| Description | Shortcut |
|---|---|
| Jump to screen | Ctrl + a + insert_screen_number |
| Return to last screen | Ctrl + a + Ctrl + a |
| Create new screen | Ctrl + a + c |
| Jump to the beginning of line | Ctrl + a + a |
| Jump to the end of line | Ctrl + e |
Quit screen |
Ctrl + d |
Using the O2 Portal
When working with big datasets, it can beneficial to have access to an IDE (intergated development environment) while writing your code. IDE’s provide a graphical user interface (GUI) such as RStudio or MatLab allowing you to view your environment and plots all within the same space.
O2 has a service called the O2 Portal which allows for users to utilize a GUI on O2. This allows you to work more conveniently on the cluster where your data is stored and access increased computational power. In order to use the O2 Portal, you need to submit a job request like you would on the command-line via the sbatch command and you will provide it SLURM directives like numbers of CPUs, memory, partition and time. Once you are queued, it will generate a session for you to use your application in. More information on the O2 Portal can be found HMS-RC’s resource page and a tutorial on how to use R Studio on the O2 Portal can be found here.
This lesson has been developed by members of the teaching team at the Harvard Chan Bioinformatics Core (HBC). These are open access materials distributed under the terms of the Creative Commons Attribution license (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.