Approximate time: 165 minutes
Learning Objectives:
- Determine how functions are attributed to genes using Gene Ontology terms
- Understand the theory of how functional enrichment tools yield statistically enriched functions or interactions
- Discuss functional analysis using over-representation analysis, functional class scoring, and pathway topology methods
- Explore functional analysis tools
Functional analysis
The output of RNA-Seq differential expression analyses, proteomic analyses, as well as other genetic/genomic analyses is a list of significant genes. To gain greater biological insight into the list of significant genes there are various analyses that can be done:
- determine whether there is enrichment of known biological functions, interactions, or pathways
- identify genes’ involvement in novel pathways or networks by grouping genes together based on similar trends
- use global changes in gene expression by visualizing all genes being significantly up- or down-regulated in the context of external interaction data
Generally for any differential expression analysis, it is useful to interpret the resulting gene lists using freely available web- and R-based tools. While tools for functional analysis span a wide variety of techniques, they can loosely be categorized into three main types: over-representation analysis, functional class scoring, and pathway topology [1].
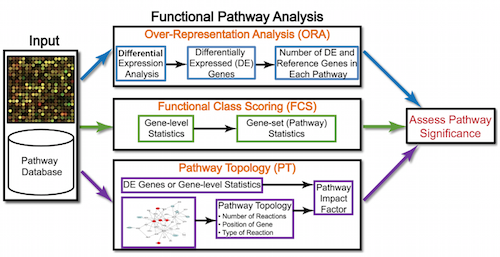
Illustration adapted from Khatri, P., Sirota, M. & Butte, A.J., PLoS Comput Biol 2012
Note: All tools described below are excellent to validate experimental results and to make hypotheses. These tools suggest genes/pathways that may be involved with your condition of interest; however, you should NOT use these tools to make conclusions about the pathways involved in your experimental process. You will need to perform experimental validation of any suggested pathways.
Over-representation analysis
The first main category of functional analysis tools is the over-representation analysis, which explores whether there is enrichment of known biological functions in a particular set of genes (e.g. significant DE genes). There are a plethora of functional enrichment tools that perform some type of over-representation analysis by querying databases containing information about gene function and interactions. Querying these databases for gene function requires the use of a controlled vocabulary to describe gene function. One of the most widely-used vocabularies is the Gene Ontology (GO). This vocabulary was established by the Gene Ontology project, and the words in the vocabulary are referred to as GO terms.
Gene Ontology project
“The Gene Ontology project is a collaborative effort to address the need for consistent descriptions of gene products across databases” [2]. The Gene Ontology Consortium maintains the GO terms, and these GO terms are incorporated into gene annotations in many of the popular repositories for animal, plant, and microbial genomes.
Tools that investigate enrichment of biological functions or interactions can query these databases for GO terms associated with a list of genes to determine whether any GO terms associated with particular functions or interactions are enriched in the gene set. Therefore, to best use and interpret the results from these functional analysis tools, it is helpful to have a good understanding of the GO terms themselves.
GO terms
GO Ontologies
To describe the roles of genes and gene products, GO terms are organized into three independent controlled vocabularies (ontologies) in a species-independent manner:
- Biological process: refers to the biological role involving the gene or gene product, and could include “transcription”, “signal transduction”, and “apoptosis”. A biological process generally involves a chemical or physical change of the starting material or input.
- Molecular function: represents the biochemical activity of the gene product, such activities could include “ligand”, “GTPase”, and “transporter”.
- Cellular component: refers to the location in the cell of the gene product. Cellular components could include “nucleus”, “lysosome”, and “plasma membrane”.
Each GO term has a term name (e.g. DNA repair) and a unique term accession number (GO:0005125), and a single gene product can be associated with many GO terms, since a single gene product “may function in several processes, contain domains that carry out diverse molecular functions, and participate in multiple alternative interactions with other proteins, organelles or locations in the cell” [3].
GO term hierarchy
Some gene products are well-researched, with vast quantities of data available regarding their biological processes and functions. However, other gene products have very little data available about their roles in the cell.
For example, the protein, “p53”, would contain a wealth of information on it’s roles in the cell, whereas another protein might only be known as a “membrane-bound protein” with no other information available.
The GO ontologies were developed to describe and query biological knowledge with differing levels of information available. To do this, GO ontologies are loosely hierarchical, ranging from general, ‘parent’, terms to more specific, ‘child’ terms. The GO ontologies are “loosely” hierarchical since ‘child’ terms can have multiple ‘parent’ terms.
Some genes with less information may only be associated with general ‘parent’ terms or no terms at all, while other genes with a lot of information be associated with many terms.
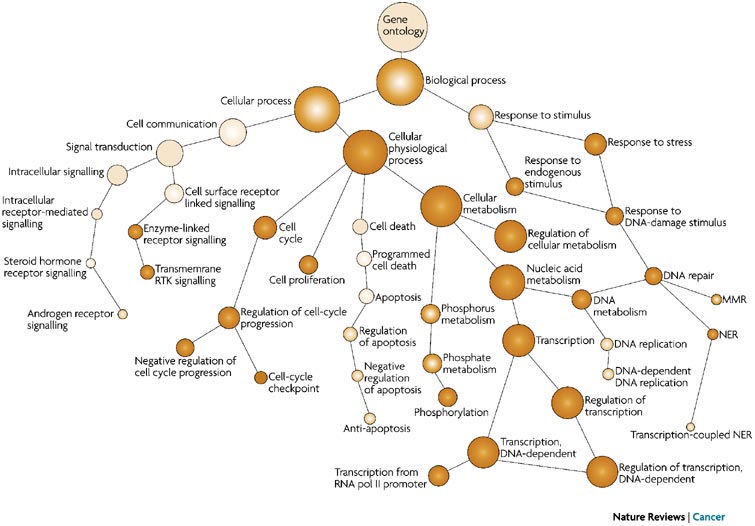
Illustration adapted from Hu P et al, Nature Reviews Cancer 2007
Tips for working with GO terms
Hypergeometric testing
So, how do we work with a database of controlled vocabularies like GO?
One of the analysis you can perform is to test if certain GO terms are enriched in you small dataset relative by determining if they occur more or less frequently than in a “background” set of genes. This can inform us about whether certain GO terms are over- or under-represented in a given gene list, and can be tested using the hypergeometric test.
This type of testing can also be utilized to test for over- or under-representation of other entities such as particular motifs or pathways as well.
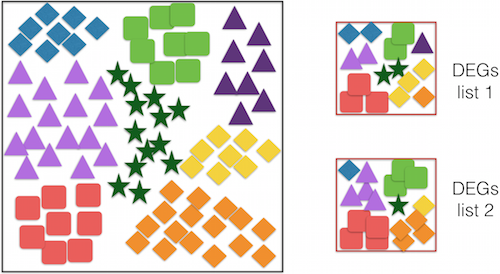
To determine whether GO terms (or motifs and pathways) are over- or under-represented, you can use the hypergeometric test to determine the probability of having the observed proportion of genes associated with a specific GO term in your gene list based on the proportion of genes associated with the same GO term in the background set. See example below:
Background set:
-
Of the 13,000 genes in the honeybee genome, 85 genes are associated with the GO term “DNA repair”.
-
Proportion of genes associated with “DNA repair” is 85/13000 which is 0.65%
Gene list of interest:
-
Your gene list of 1,000 genes has 50 genes associated with “DNA repair”.
-
Proportion of genes associated with “DNA repair” is 50/1000 which is 5%
In this example it is evident that the GO term “DNA repair” is over-represented in the gene list of interest relative to the background set.
The “background gene set” can be all genes from an organism, or a selected subset.
To determine whether a GO term or pathway is significantly over- or under-represented, tools often perform hypergeometric testing. Using our honeybee example, the hypergeometric distribution is a probability distribution that describes the probability of 50 genes (k) being associated with “DNA repair”, for all genes in our gene list (n=1,000), from a population of all of the genes in entire genome (N=13,000) which contains 85 genes (K) associated with “DNA repair” [4].
The calculation of probability of k successes follows the formula:
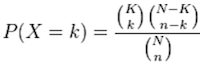
clusterProfiler
clusterProfiler performs over-representation analysis on GO terms associated with a list of genes. The tool takes as input a significant gene list and a background gene list and performs statistical enrichment analysis using hypergeometric testing. The basic arguments allow the user to select the appropriate organism and GO ontology (BP, CC, MF) to test.
Running clusterProfiler
The data we will be working with is the differential expression results for samples overexpressing the MOV10 gene versus control samples, as we discussed earlier. Based on the authors’ hypothesis, we may expect the enrichment of processes/pathways related to translation, splicing, and the regulation of mRNAs.
Let’s get started with the res_tableOE dataframe we created during set up.
To run clusterProfiler GO over-representation analysis, we will change our gene names into Ensembl IDs, since the tool works a bit easier with the Ensembl IDs. There are a few clusterProfiler functions that allow us to map between gene IDs:
## clusterProfiler does not work as easily using gene names, so we will turn gene names into Ensembl IDs using
## clusterProfiler::bitr and merge the IDs back with the DE results
keytypes(org.Hs.eg.db)
ids <- bitr(rownames(res_tableOE),
fromType = "SYMBOL",
toType = c("ENSEMBL", "ENTREZID"),
OrgDb = "org.Hs.eg.db")
## The gene names can map to more than one Ensembl ID (some genes change ID over time),
## so we need to remove duplicate IDs prior to assessing enriched GO terms
non_duplicates <- which(duplicated(ids$SYMBOL) == FALSE)
ids <- ids[non_duplicates, ]
## Merge the Ensembl IDs with the results
merged_gene_ids <- merge(x=res_tableOE, y=ids, by.x="row.names", by.y="SYMBOL")
## Extract significant results
sigOE <- subset(merged_gene_ids, padj < 0.05)
sigOE_genes <- as.character(sigOE$ENSEMBL)
We will use the Ensembl IDs for all genes as the background dataset:
## Create background dataset for hypergeometric testing using all genes tested for significance in the results
allOE_genes <- as.character(merged_gene_ids$ENSEMBL)
Now we can perform the GO enrichment analysis:
## Run GO enrichment analysis
ego <- enrichGO(gene = sigOE_genes,
universe = allOE_genes,
keytype = "ENSEMBL",
OrgDb = org.Hs.eg.db,
ont = "BP",
pAdjustMethod = "BH",
qvalueCutoff = 0.05,
readable = TRUE)
NOTE: The library used for the annotations associated with genes (here we are using
org.Hs.eg.db) will change based on organism (e.g. if studying mouse, would need to install and loadorg.Mm.eg.db). The list of different organism packages are given here.
Output results to a data frame and save to file:
## Output results from GO analysis to a table
cluster_summary <- data.frame(ego)
write.csv(cluster_summary, "results/clusterProfiler_Mov10oe.csv")
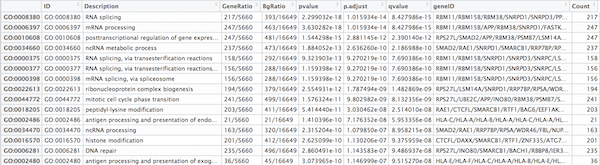
Exercises
-
Using the same conditions, return the enriched GO processes for the Molecular Function ontology as
egoMF. -
How would the
enrichGO()function change if our organism was rat?
Visualizing clusterProfiler results
clusterProfiler has a variety of options for viewing the over-represented GO terms. We will explore the dotplot, enrichment plot, and the category netplot.
The dotplot shows the number of genes associated with the first 50 terms (size) and the p-adjusted values for these terms (color).
## Dotplot gives top 50 genes by gene ratio (# genes related to GO term / total number of sig genes), not padj.
dotplot(ego, showCategory=50)
To save the figure, click on the Export button in the RStudio Plots tab and Save as PDF.... In the pop-up window, change:
Orientation:toLandscapePDF sizeto8 x 14to give a figure of appropriate size for the text labels
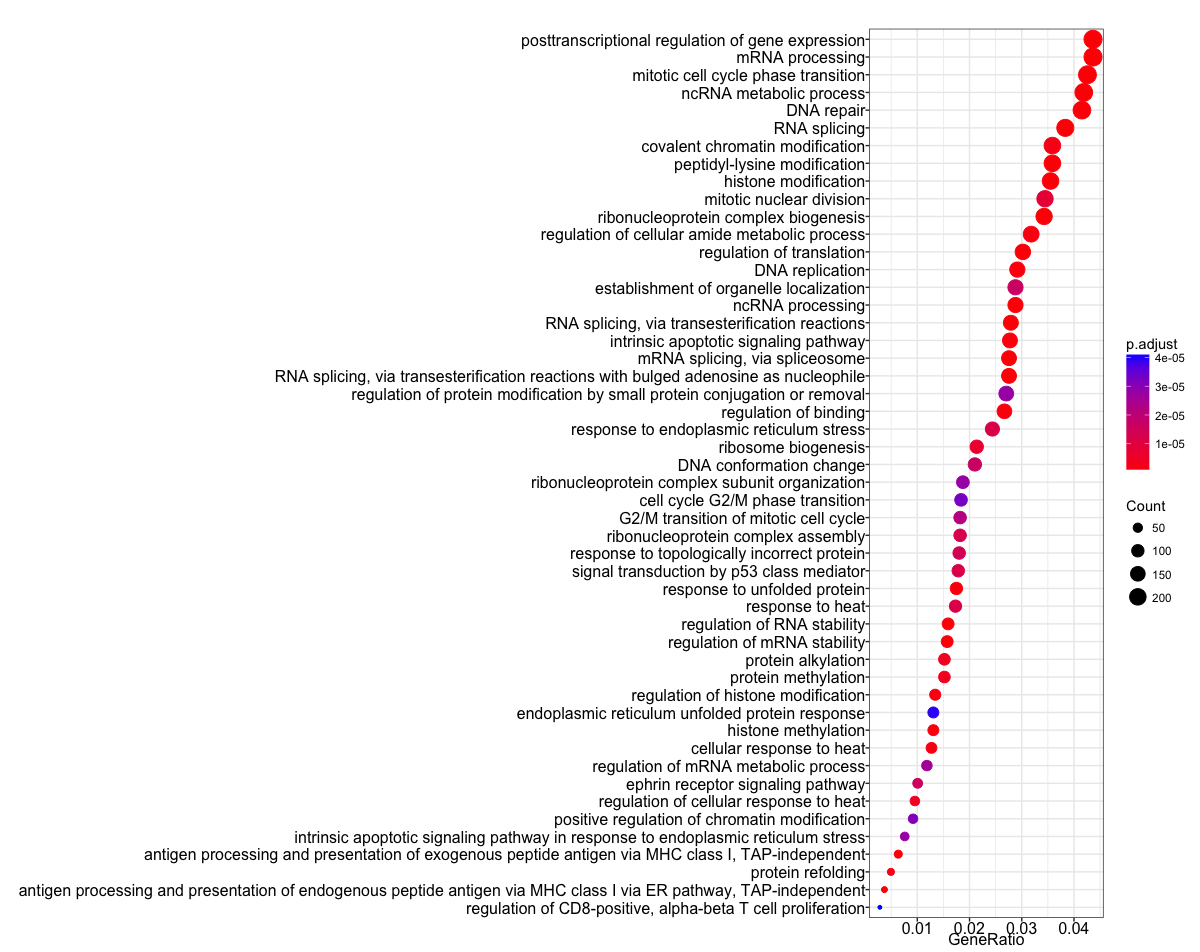
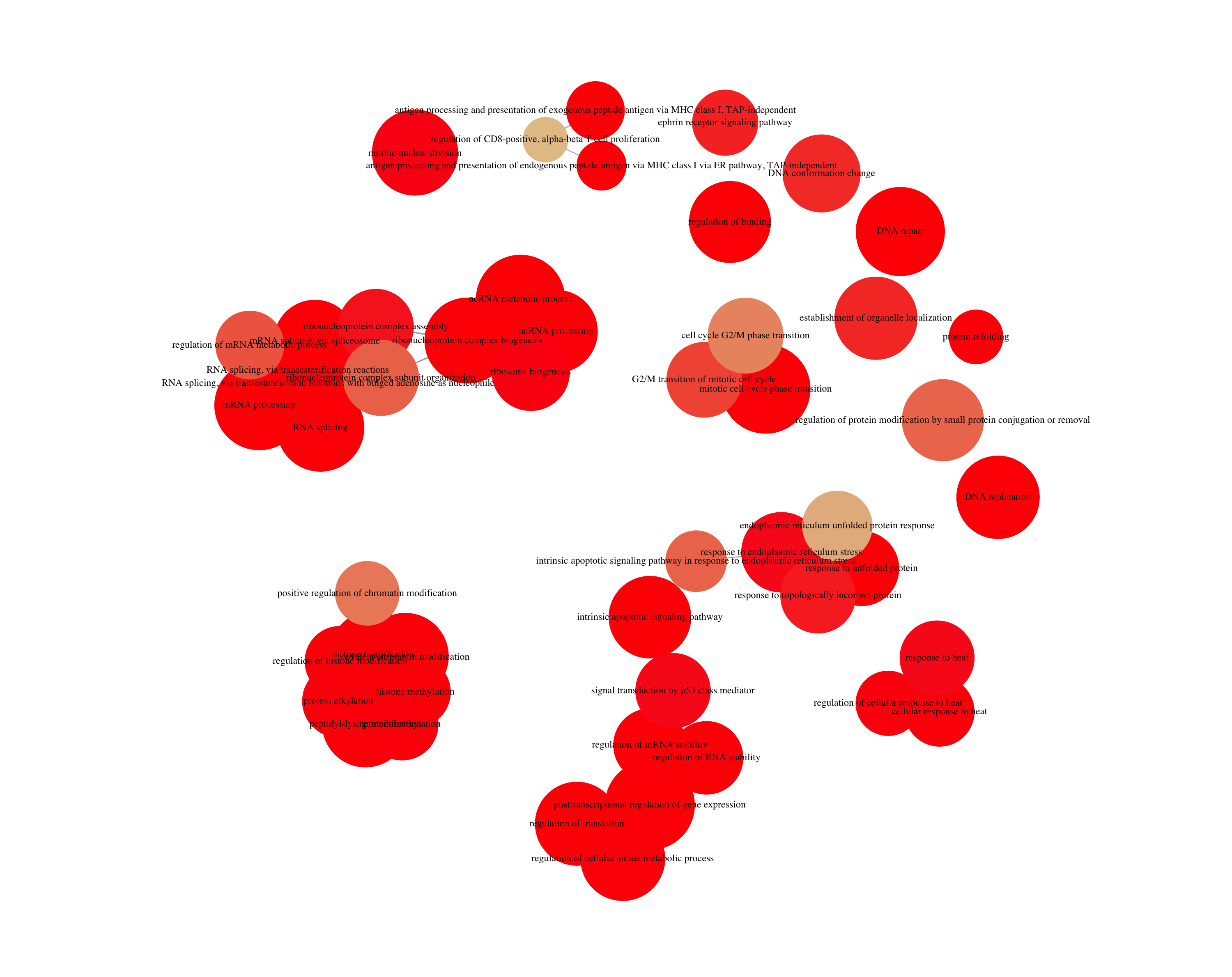
The next plot is the enrichment GO plot, which shows the relationship between the top 50 most significantly enriched GO terms, by grouping similar terms together. The color represents the p-values relative to the other displayed terms (brighter red is more significant) and the size of the terms represents the number of genes that are significant from our list.
## Enrichmap clusters the 50 most significant (by padj) GO terms to visualize relationships between terms
enrichMap(ego, n=50, vertex.label.font=6)
To save the figure, click on the Export button in the RStudio Plots tab and Save as PDF.... In the pop-up window, change the PDF size to 24 x 32 to give a figure of appropriate size for the text labels.
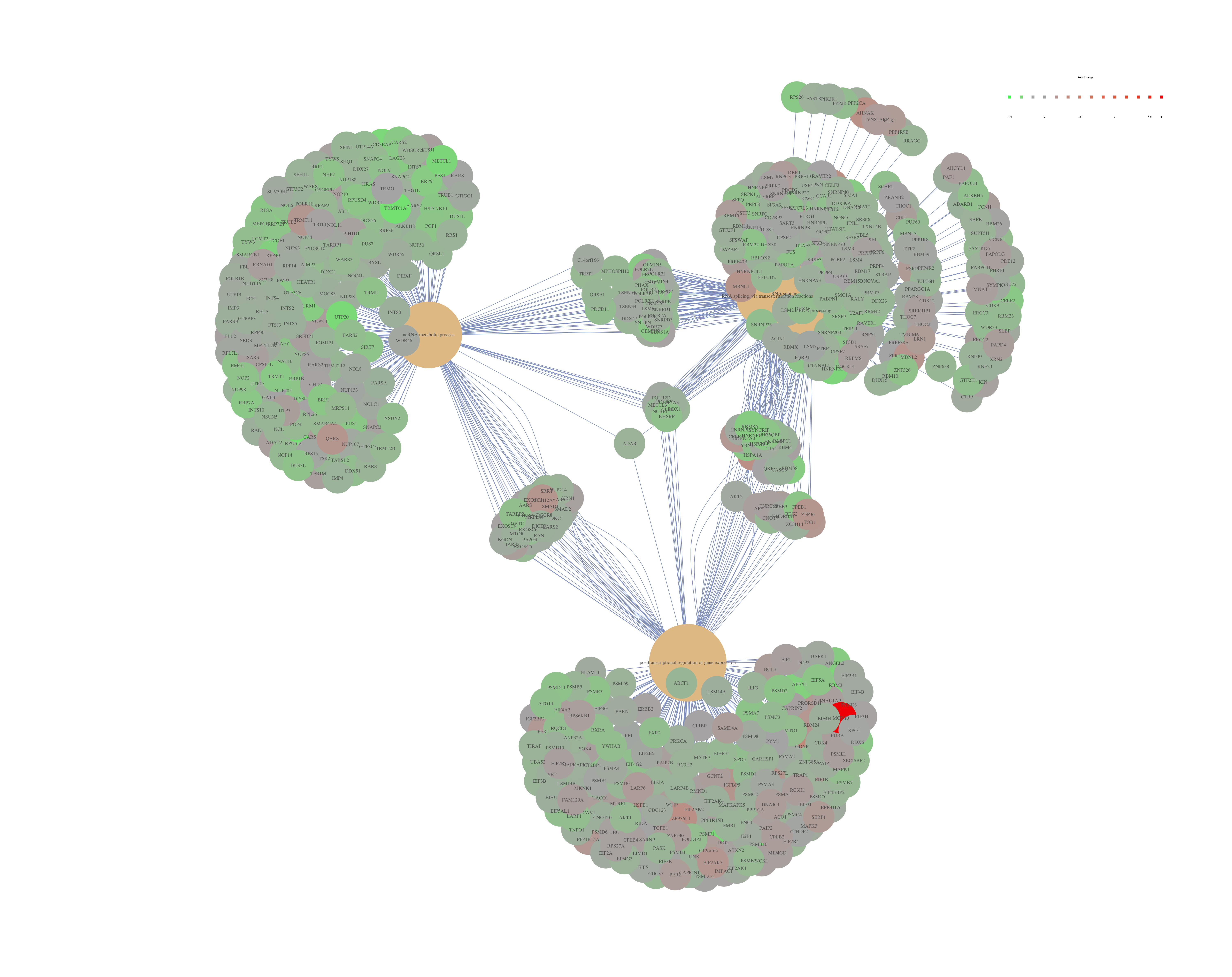
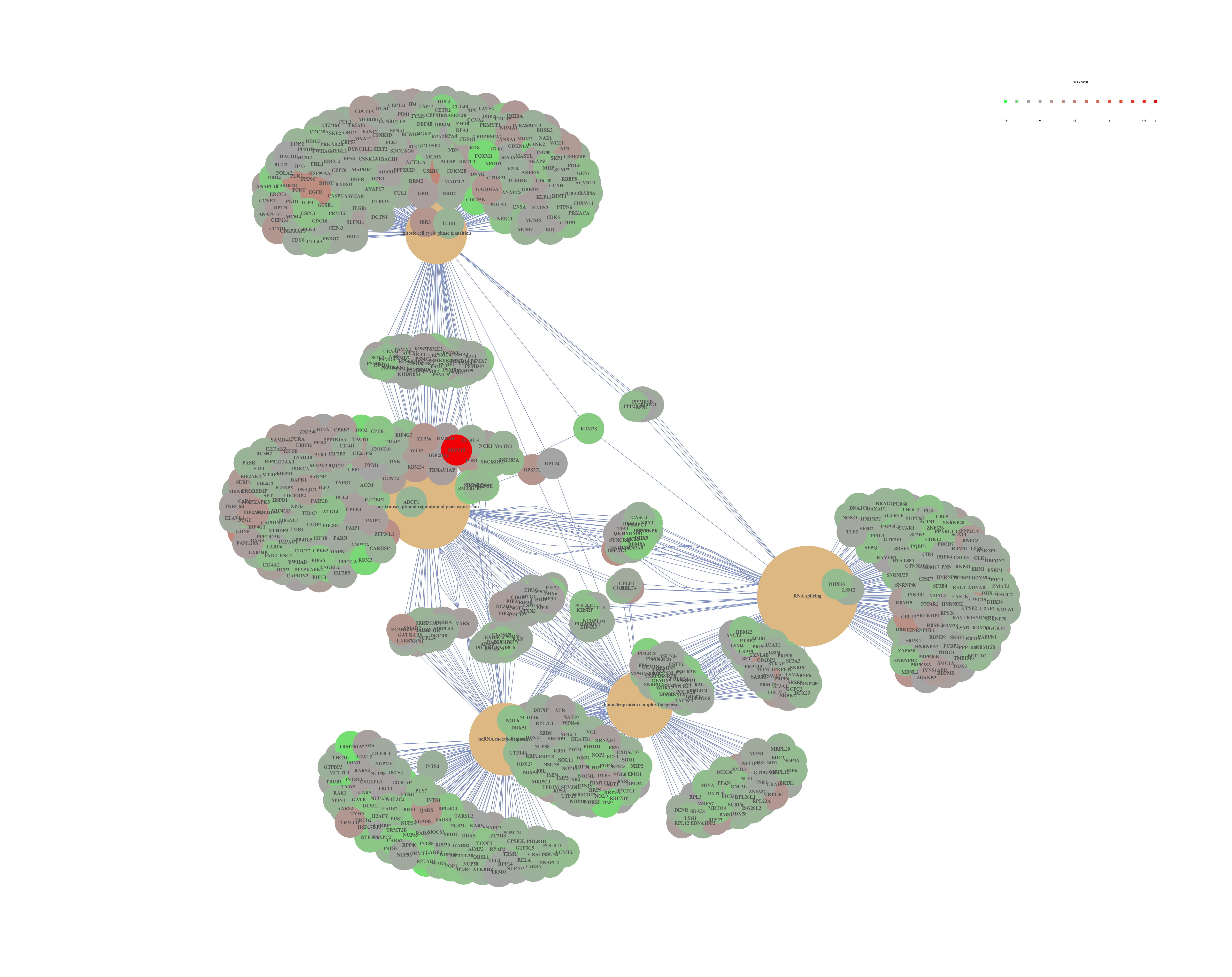
Finally, the category netplot shows the relationships between the genes associated with the top five most significant GO terms and the fold changes of the significant genes associated with these terms (color). The size of the GO terms reflects the pvalues of the terms, with the more significant terms being larger. This plot is particularly useful for hypothesis generation in identifying genes that may be important to several of the most affected processes.
## To color genes by log2 fold changes, we need to extract the log2 fold changes from our results table creating a named vector
OE_foldchanges <- sigOE$log2FoldChange
names(OE_foldchanges) <- sigOE$Row.names
## Cnetplot details the genes associated with one or more terms - by default gives the top 5 significant terms (by padj)
cnetplot(ego,
categorySize="pvalue",
showCategory = 5,
foldChange=OE_foldchanges,
vertex.label.font=6)
## If some of the high fold changes are getting drowned out due to a large range, you could set a minimum and maximum fold change value
OE_foldchanges <- ifelse(OE_foldchanges > 2, 2, OE_foldchanges)
OE_foldchanges <- ifelse(OE_foldchanges < -2, -2, OE_foldchanges)
cnetplot(ego,
categorySize="pvalue",
showCategory = 5,
foldChange=OE_foldchanges,
vertex.label.font=6)
Again, to save the figure, click on the Export button in the RStudio Plots tab and Save as PDF.... Change the PDF size to 48 x 56 to give a figure of appropriate size for the text labels.
If you are interested in significant processes that are not among the top five, you can subset your ego dataset to only display these processes:
## Subsetting the ego results without overwriting original ego variable
ego2 <- ego
ego2@result <- ego@result[c(1,3,4,8,9),]
## Plotting terms of interest
cnetplot(ego2, categorySize="pvalue", foldChange=OE_foldchanges, showCategory = 5)
Exercises
-
Create a dotplot for the
egoMFresults and show only the top 25 processes by gene ratio. -
Create an enrichment GO plot for the
egoMFresults and show only the top 25 most significant processes. -
Create a category netplot for the
egoMFresults using only the following GO ids:
"GO:0043021", "GO:0003729", "GO:0003730", "GO:0004386", "GO:0045296"
gProfiler
gProfileR is another tool for performing ORA, similar to clusterProfiler. gProfileR considers multiple sources of functional evidence, including Gene Ontology terms, biological pathways, regulatory motifs of transcription factors and microRNAs, human disease annotations and protein-protein interactions. The user selects the organism and the sources of evidence to test. There are also additional parameters to change various thresholds and tweak the stringency to the desired level.
The GO terms output by gprofileR are generally quite similar to those output by clusterProfiler, but there are small differences due to the different algorithms used by the tools. We often use gProfiler with REVIGO to collapse redundant GO terms by semantic similarity and summarize them graphically. If you are interested in exploring gProfileR/REVIGO on your own, we have provided the instructional materials.
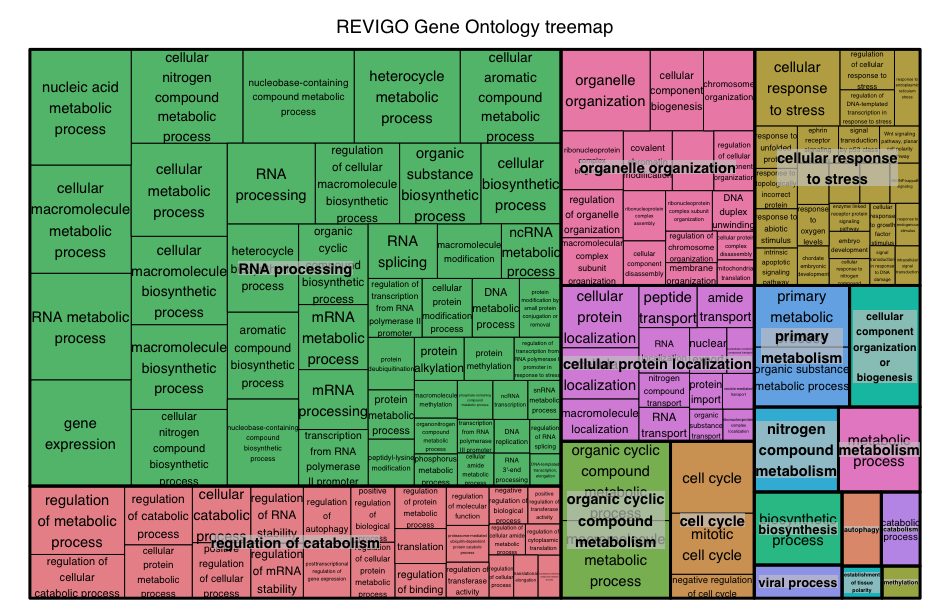
Over-representation analysis is only a single type of functional analysis method that is available for teasing apart the biological processes important to your condition of interest. Other types of analyses can be equally important or informative, including functional class scoring and pathway topology methods.
Functional class scoring tools
Functional class scoring (FCS) tools, such as GSEA, most often use the gene-level statistics or log2 fold changes for all genes from the differential expression results, then look to see whether gene sets for particular biological pathways are enriched among the large positive or negative fold changes.
The hypothesis of FCS methods is that although large changes in individual genes can have significant effects on pathways (and will be detected via ORA methods), weaker but coordinated changes in sets of functionally related genes (i.e., pathways) can also have significant effects. Thus, rather than setting an arbitrary threshold to identify ‘significant genes’, all genes are considered in the analysis. The gene-level statistics from the dataset are aggregated to generate a single pathway-level statistic and statistical significance of each pathway is reported. This type of analysis can be particularly helpful if the differential expression analysis only outputs a small list of significant DE genes.
Gene set enrichment analysis using clusterProfiler and Pathview
Using the log2 fold changes obtained from the differential expression analysis for every gene, gene set enrichment analysis and pathway analysis can be performed using clusterProfiler and Pathview tools.
For gene set or pathway analysis using clusterProfiler, coordinated differential expression over gene sets is tested instead of changes of individual genes. “Gene sets are pre-defined groups of genes, which are functionally related. Commonly used gene sets include those derived from KEGG pathways, Gene Ontology terms, MSigDB, Reactome, or gene groups that share some other functional annotations, etc. Consistent perturbations over such gene sets frequently suggest mechanistic changes” [1].
To perform GSEA analysis of KEGG gene sets, clusterProfiler requires the genes to be identified using Entrez IDs for all genes in our results dataset. We also need to remove the NA values and duplicates (due to gene ID conversion) prior to the analysis:
## Remove any NA values
all_results_gsea <- subset(merged_gene_ids, ENTREZID != "NA")
## Remove any duplicates
all_results_gsea <- all_results_gsea[which(duplicated(all_results_gsea$Row.names) == F), ]
Finally, extract and name the fold changes:
## Extract the foldchanges
foldchanges <- all_results_gsea$log2FoldChange
## Name each fold change with the corresponding Entrez ID
names(foldchanges) <- all_results_gsea$ENTREZID
Order the foldchanges in decreasing order:
## Sort fold changes in decreasing order
foldchanges <- sort(foldchanges, decreasing = TRUE)
head(foldchanges)
Perform the GSEA using KEGG gene sets:
## GSEA using gene sets from KEGG pathways
gseaKEGG <- gseKEGG(geneList = foldchanges, # ordered named vector of fold changes (Entrez IDs are the associated names)
organism = "hsa",
nPerm = 1000, # default number permutations
minGSSize = 120, # minimum gene set size (# genes in set) - change to test more sets or recover sets with fewer # genes
pvalueCutoff = 0.05, # padj cutoff value
verbose = FALSE)
## Extract the GSEA results
gseaKEGG_results <- gseaKEGG@result
NOTE: The organisms with KEGG pathway information are listed here.
View the enriched pathways:
## Write GSEA results to file
write.csv(gseaKEGG_results, "results/gseaOE.csv", quote=F)
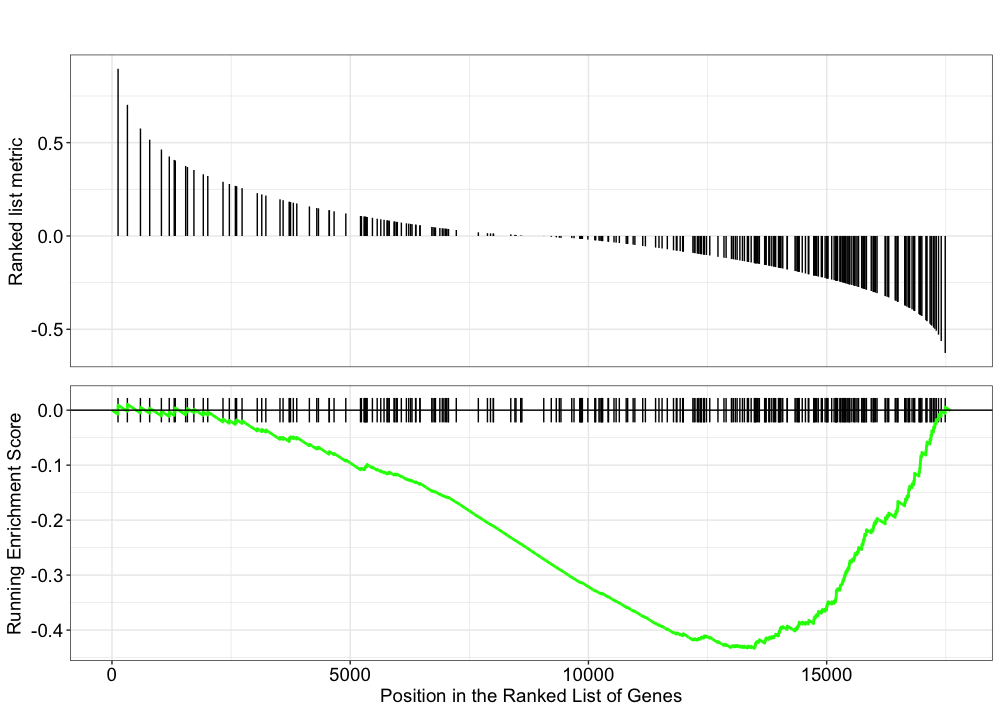
Explore the GSEA plot of enrichment of the pathway-associated genes in the ranked list:
## Plot the GSEA plot for a single enriched pathway, `hsa03040`
gseaplot(gseaKEGG, geneSetID = 'hsa03040')
Use the Pathview R package to integrate the KEGG pathway data from clusterProfiler into pathway images:
## Output images for a single significant KEGG pathway
pathview(gene.data = foldchanges,
pathway.id = "hsa03040",
species = "hsa",
limit = list(gene = 2, # value gives the max/min limit for foldchanges
cpd = 1))
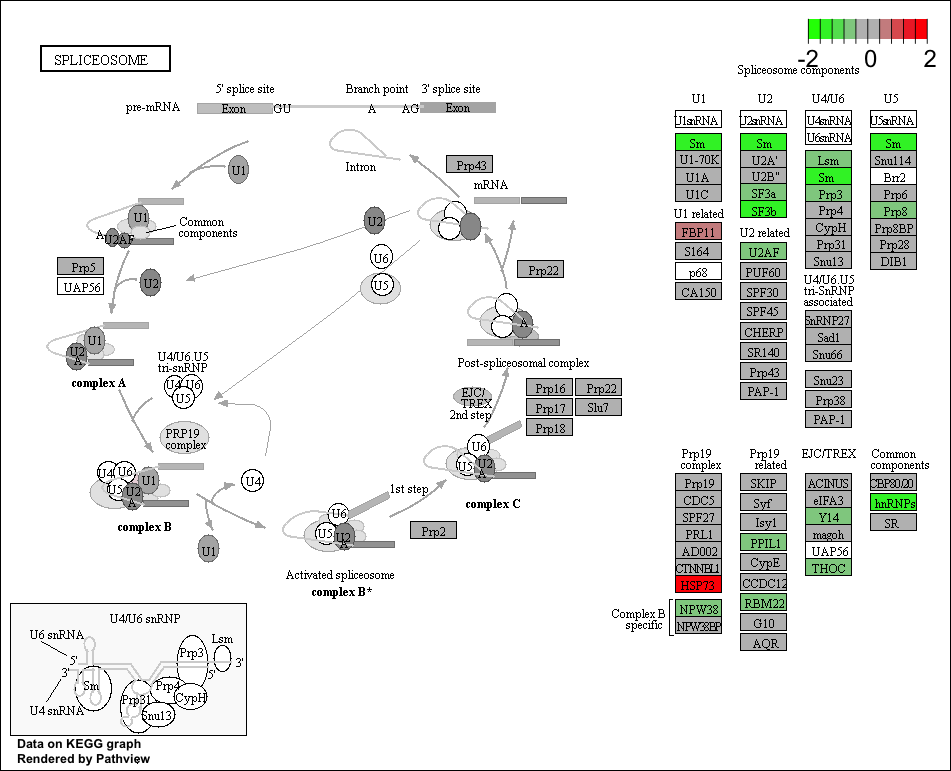
NOTE: Printing out Pathview images for all significant pathways can be easily performed as follows:
## Output images for all significant KEGG pathways get_kegg_plots <- function(x) { pathview(gene.data = foldchanges, pathway.id = gseaKEGG_results$ID[x], species = "hsa", limit = list(gene = 2, cpd = 1)) } library(purrr) map(1:length(gseaKEGG_results$ID), get_kegg_plots)
Exercise
You are interested in some pathways that are comprised of only ~50 genes, how can you modify the gseaKEGG() to include these pathways for enrichment testing? How would you change the function if your organism was rat?
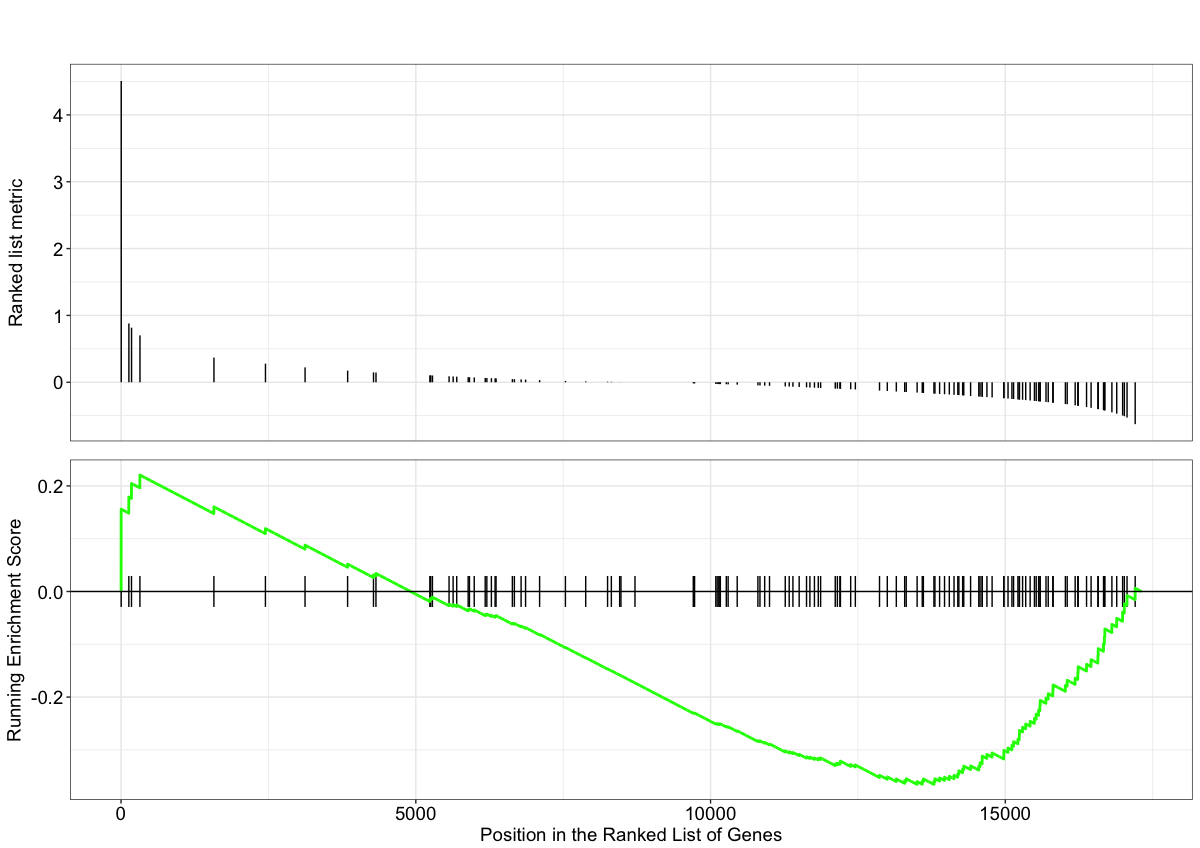
Instead of exploring enrichment of KEGG gene sets, we can also explore the enrichment of BP Gene Ontology terms using gene set enrichment analysis:
# GSEA using gene sets associated with BP Gene Ontology terms
gseaGO <- gseGO(geneList = foldchanges,
OrgDb = org.Hs.eg.db,
ont = 'BP',
nPerm = 1000,
minGSSize = 120,
pvalueCutoff = 0.05,
verbose = FALSE)
gseaGO_results <- gseaGO@result
gseaplot(gseaGO, geneSetID = 'GO:0007423')
There are other gene sets available for GSEA analysis in clusterProfiler (Disease Ontology, Reactome pathways, etc.). In addition, it is possible to supply your own gene set GMT file, such as a GMT for MSigDB downloaded from the Broad Institute website using special clusterProfiler functions as shown below:
# DO NOT RUN
biocLite("GSEABase")
library(GSEABase)
# Load in GMT file of gene sets (we downloaded MSigDB GMT from Broad)
c2 <- read.gmt("/data/c2.cp.v6.0.entrez.gmt.txt")
msig <- GSEA(foldchanges, TERM2GENE=c2, verbose=FALSE)
msig_df <- data.frame(msig)
Pathway topology tools
The last main type of functional analysis technique is pathway topology analysis. Pathway topology analysis often takes into account gene interaction information along with the fold changes and adjusted p-values from differential expression analysis to identify dysregulated pathways. Depending on the tool, pathway topology tools explore how genes interact with each other (e.g. activation, inhibition, phosphorylation, ubiquitination, etc.) to determine the pathway-level statistics. Pathway topology-based methods utilize the number and type of interactions between gene product (our DE genes) and other gene products to infer gene function or pathway association.
SPIA
The SPIA (Signaling Pathway Impact Analysis) tool can be used to integrate the lists of differentially expressed genes, their fold changes, and pathway topology to identify affected pathways. The blog post from Getting Genetics Done provides a step-by-step procedure for using and understanding SPIA.
# Set-up
## Significant genes is a vector of fold changes where the names are ENTREZ gene IDs. The background set is a vector of all the genes represented on the platform.
## Convert ensembl to entrez ids
sig_genes_entrez <- subset(all_results_gsea, padj< 0.05)
sig_genes <- sig_genes_entrez$log2FoldChange
names(sig_genes) <- sig_genes_entrez$ENTREZID
head(sig_genes)
## Remove NA and duplicated values (if not already removed)
sig_genes <- sig_genes[!is.na(names(sig_genes))]
sig_genes <- sig_genes[!duplicated(names(sig_genes))]
background_genes <- merged_gene_ids$ENTREZID
background_genes <- background_genes[!is.na(background_genes)]
background_genes <- background_genes[!duplicated(background_genes)]
Now that we have our background and significant genes in the appropriate format, we can run SPIA:
## Run SPIA.
spia_result <- spia(de=sig_genes, all=background_genes, organism="hsa")
head(spia_result, n=20)
SPIA outputs a table showing significantly dysregulated pathways based on over-representation and signaling perturbations accumulation. The table shows the following information:
pSize: the number of genes on the pathwayNDE: the number of DE genes per pathwaytA: the observed total perturbation accumulation in the pathwaypNDE: the probability to observe at least NDE genes on the pathway using a hypergeometric model (similar to ORA)pPERT: the probability to observe a total accumulation more extreme than tA only by chancepG: the p-value obtained by combining pNDE and pPERTpGFdrandpGFWERare the False Discovery Rate and respectively Bonferroni adjusted global p-valuesStatus: gives the direction in which the pathway is perturbed (activated or inhibited)KEGGLINKgives a web link to the KEGG website that displays the pathway image with the differentially expressed genes highlighted in red
We can view the significantly dysregulated pathways by viewing the over-representation and perturbations for each pathway.
plotP(spia_result, threshold=0.05)
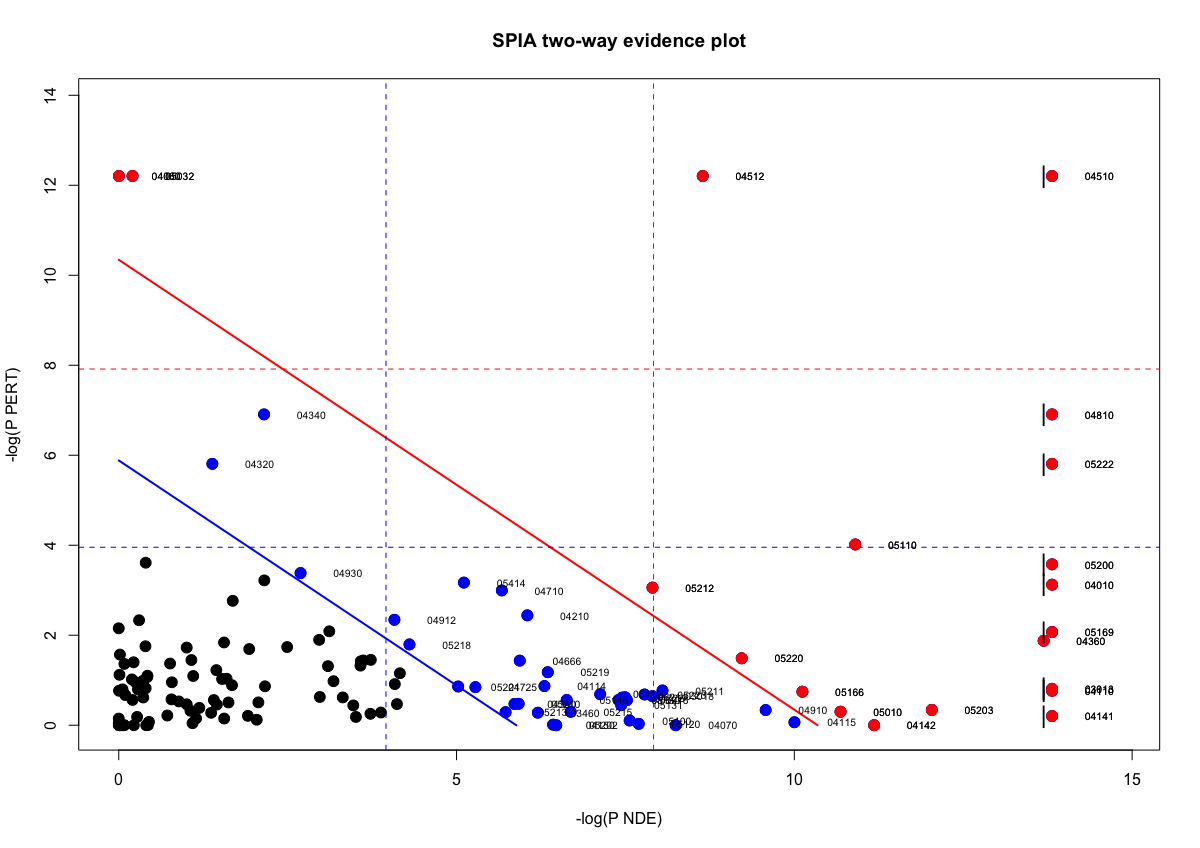
In this plot, each pathway is a point and the coordinates are the log of pNDE (using a hypergeometric model) and the p-value from perturbations, pPERT. The oblique lines in the plot show the significance regions based on the combined evidence.
If we choose to explore the significant genes from our dataset occurring in these pathways, we can subset our SPIA results:
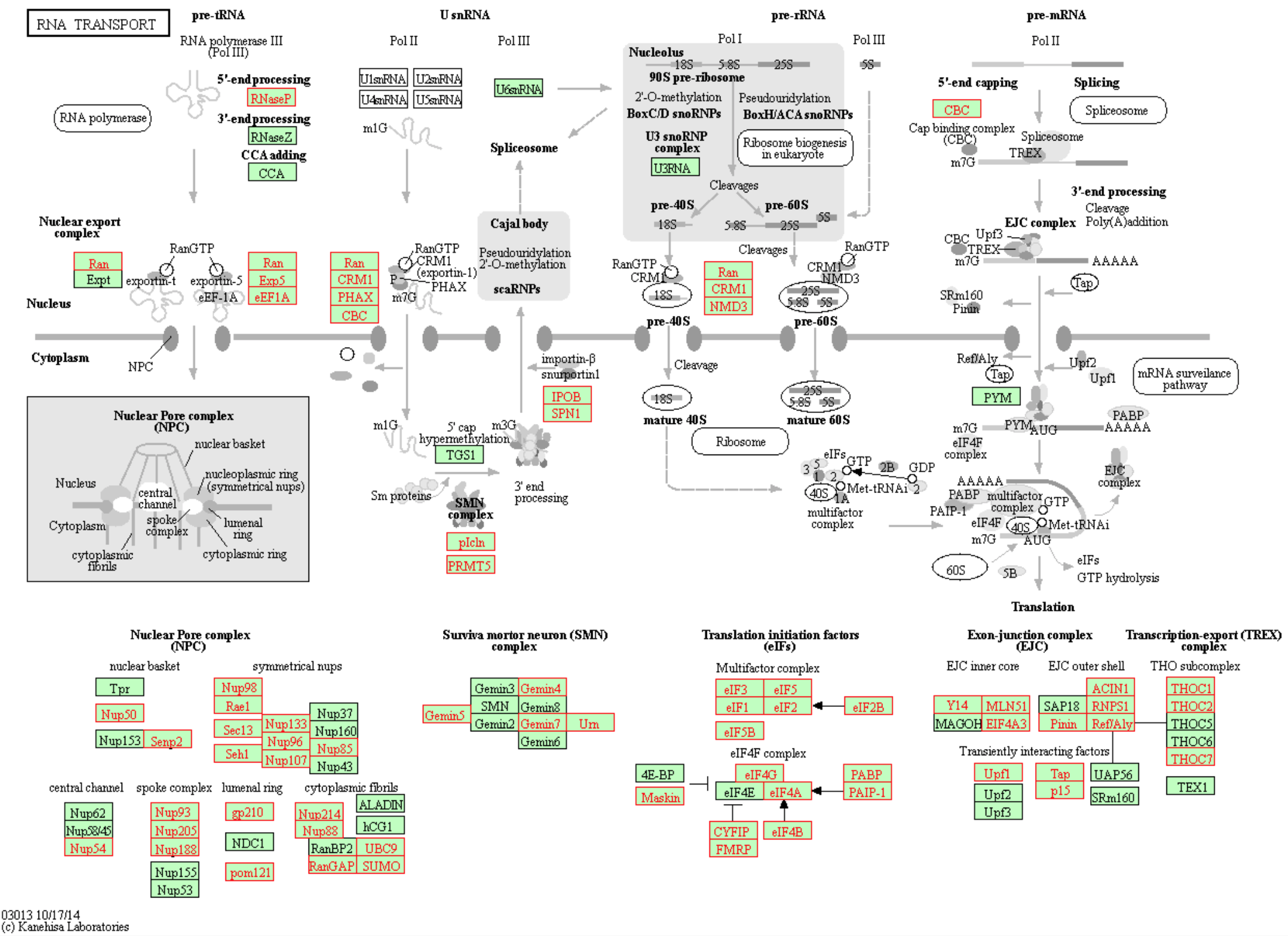
## Look at pathway 03013 and view kegglink
subset(spia_result, ID == "03013")
Copy the link KEGGLINK output on the console into a web browser to view the genes within our dataset from these perturbed pathways:
Exercise
Let’s suppose you want to focus on pathways with genes enriched within the list of significant genes compared to the background AND highly perturbing to the pathway. Based on the spia plot, which pathways might you explore?
Other Tools
GeneMANIA
GeneMANIA is a tool for predicting the function of your genes. Rather than looking for enrichment, the query gene set is evaluated in the context of curated functional association data and results are displayed in the form of a network. Association data include protein and genetic interactions, pathways, co-expression, co-localization and protein domain similarity. Genes are represented as the nodes of the network and edges are formed by known association evidence. The query gene set is highlighted and so you can find other genes that are related based on the toplogy in the network. This tool is more useful for smaller gene sets (< 400 genes), as you can see in the figure below our input results in a bit of a hairball that is hard to interpret.
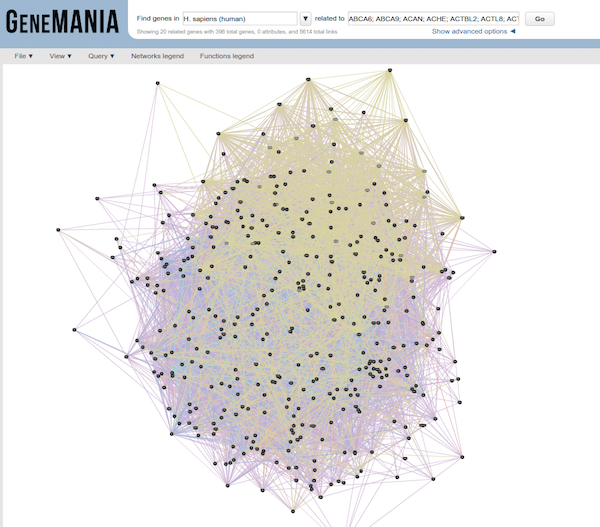
Use the significant gene list generated from the analysis we performed in class as input to GeneMANIA. Using only pathway and coexpression data as evidence, take a look at the network that results. Can you predict anything functionally from this set of genes?
Co-expression clustering
Co-expression clustering is often used to identify genes of novel pathways or networks by grouping genes together based on similar trends in expression. These tools are useful in identifying genes in a pathway, when their participation in a pathway and/or the pathway itself is unknown. These tools cluster genes with similar expression patterns to create ‘modules’ of co-expressed genes which often reflect functionally similar groups of genes. These ‘modules’ can then be compared across conditions or in a time-course experiment to identify any biologically relevant pathway or network information.
You can visualize co-expression clustering using heatmaps, which should be viewed as suggestive only; serious classification of genes needs better methods.
The way the tools perform clustering is by taking the entire expression matrix and computing pair-wise co-expression values. A network is then generated from which we explore the topology to make inferences on gene co-regulation. The WGCNA package (in R) is one example of a more sophisticated method for co-expression clustering.

Resources for functional analysis
- g:Profiler - http://biit.cs.ut.ee/gprofiler/index.cgi
- DAVID - http://david.abcc.ncifcrf.gov/tools.jsp
- clusterProfiler - http://bioconductor.org/packages/release/bioc/html/clusterProfiler.html
- GeneMANIA - http://www.genemania.org/
- GenePattern - http://www.broadinstitute.org/cancer/software/genepattern/ (need to register)
- WebGestalt - http://bioinfo.vanderbilt.edu/webgestalt/ (need to register)
- AmiGO - http://amigo.geneontology.org/amigo
- ReviGO (visualizing GO analysis, input is GO terms) - http://revigo.irb.hr/
- WGCNA - http://www.genetics.ucla.edu/labs/horvath/CoexpressionNetwork
- GSEA - http://software.broadinstitute.org/gsea/index.jsp
- SPIA - https://www.bioconductor.org/packages/release/bioc/html/SPIA.html
- GAGE/Pathview - http://www.bioconductor.org/packages/release/bioc/html/gage.html
This lesson has been developed by members of the teaching team at the Harvard Chan Bioinformatics Core (HBC). These are open access materials distributed under the terms of the Creative Commons Attribution license (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.