Working in an HPC environment
Learning Objectives
- Define the basic components of a computing environment
- Explain parallelization in computing
- Log in to the O2 cluster and start an interactive session
- Describe the salient features of the SLURM job scheduling system
- Describe the usage of the LMOD system
- Discuss storage options on the cluster
Clarifying some terminology
Cores
CPUs (Central Processing Units) are composed of a single or multiple cores. If I have a processor with 4 “cores”, i.e. a “quad-core” processor. This means that my processor can do 4 distinct computations at the same time.
Data Storage
Data storage is measured in bytes, usually Gibibytes (GiB), Tebibytes (TiB), Pebibytes (PiB). My laptop has 500 GiB of storage, which is pretty standard for laptops.
Note: You might be confused by these units. Generally speaking, you can think of a KiB =~ kB, MiB =~ MB, GiB =~ GB and TiB =~ TB. This nonmenclature comes from the difference that computers measure space in binary (base 2), while the prefixes are derived from a metric (base 10) system. So, a KiB is actually 1024 bytes worth of space, while a KB is 1000 bytes worth of space. The table below can help further demonstrate these differences.
Unit Size in Bytes Unit Size in Bytes Kilobyte (kB) 1,0001 = 1,000 Kibibyte (KiB) 1,0241 = 1,024 Megabyte (MB) 1,0002 = 1,000,000 Mebibyte (MiB) 1,0242 = 1,048,576 Gigabyte (GB) 1,0003 = 1,000,000,000 Gibibyte (GiB) 1,0243 = 1,073,741,824 Terabyte (TB) 1,0004 = 1,000,000,000,000 Tebibyte (TiB) 1,0244 = 1,099,511,627,776 Petabyte (PB) 1,0005 = 1,000,000,000,000,000 Pebibyte (TiB) 1,0245 = 1,125,899,906,842,620
Memory
Memory is a small amount of volatile or temporary information storage. This is distinct from data storage we discussed in the previous point. It is an essential component of any computer, as this is where data is stored when some computation is being performed on it. If you have ever used R, all the objects in your environment are usually stored in memory until you save your environment. My laptop has 16 GiB of memory, which is a little higher than average.
Why use the cluster or an HPC environment?
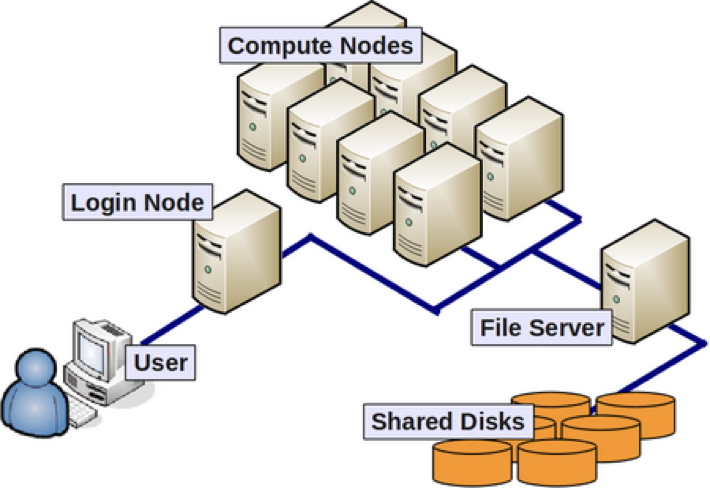
- A lot of software is designed to work with the resources on an HPC environment and is either unavailable for, or unusable on, a personal computer.
- If you are performing analysis on large data files (e.g. high-throughput sequencing data), you should work on the cluster to avoid issues with memory and to get the analysis done a lot faster with the superior processing capacity. Essentially, a cluster has:
- 100s of cores for processing!
- 100s of Gibibytes or Petabytes of storage!
- 100s of Gibibytes of memory!
Parallelization
Point 2 in the last section brings us to the idea of parallelization or parallel computing that enables us to efficiently use the resources available on the cluster.
What if we had 3 input files that we wanted to analyze? Well, we could process these files in serial, i.e. use the same core(s) over and over again, with or without multithreading, as shown in the image below.
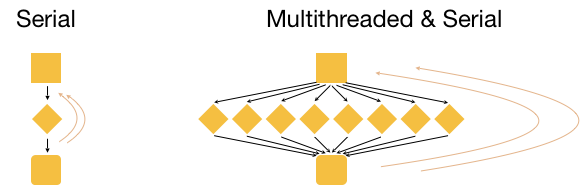
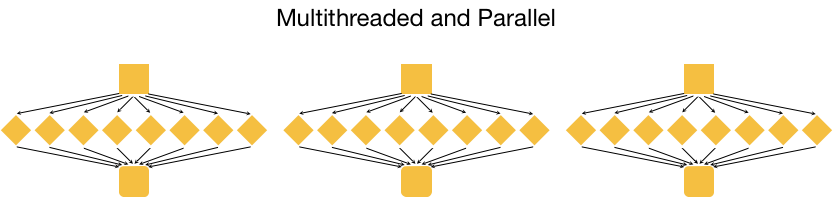
This is great, but it is not as efficient as multithreading each analysis, and using a set of 8 cores for each of the three input samples. With this type of parallelization, several samples can be analysed at the same time!
Connect to a login node on O2
Let’s get started with the hands-on component by typing in the following command to log in to O2:
ssh username@o2.hms.harvard.edu
You will receive a prompt for your password, and you should type in your associated password; note that the cursor will not move as you type in your password.
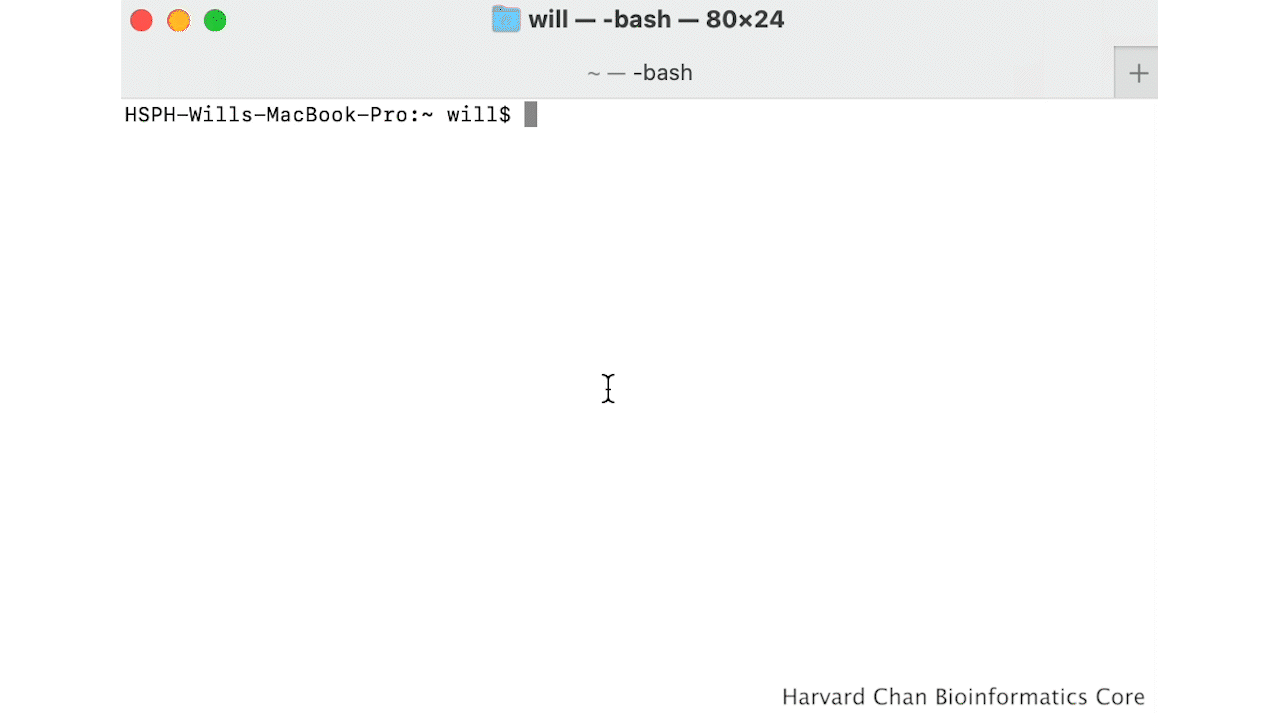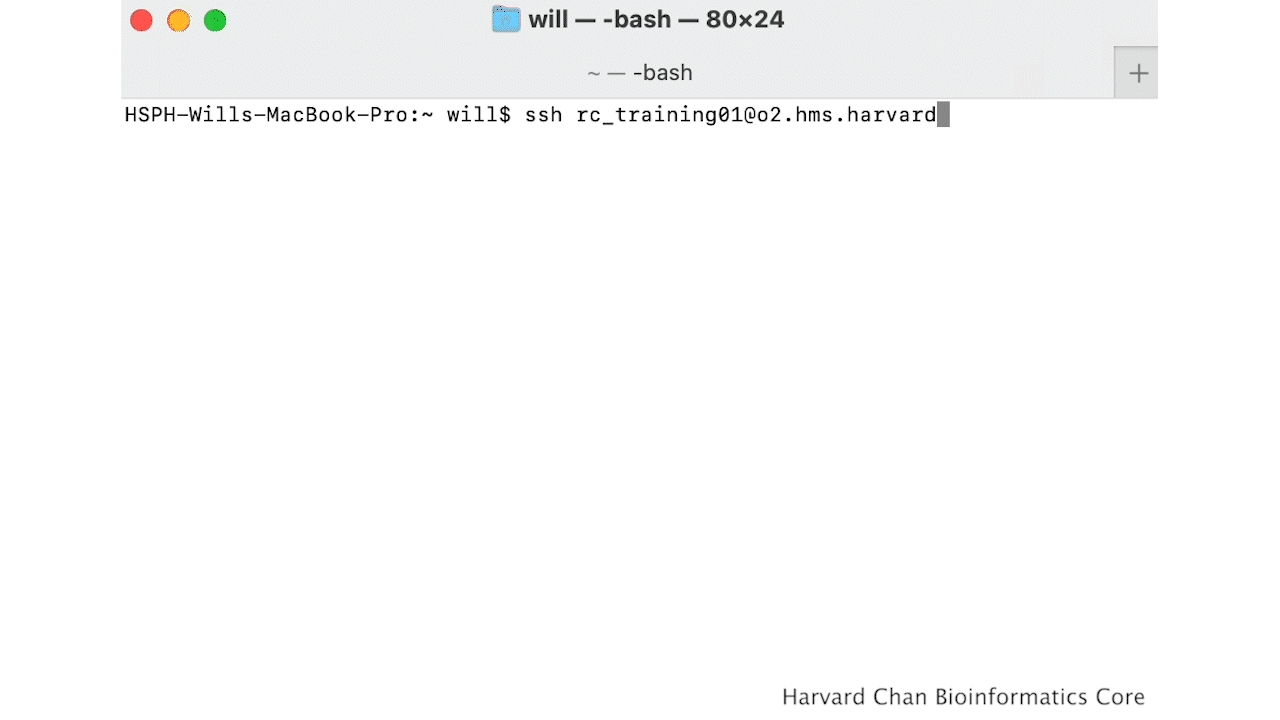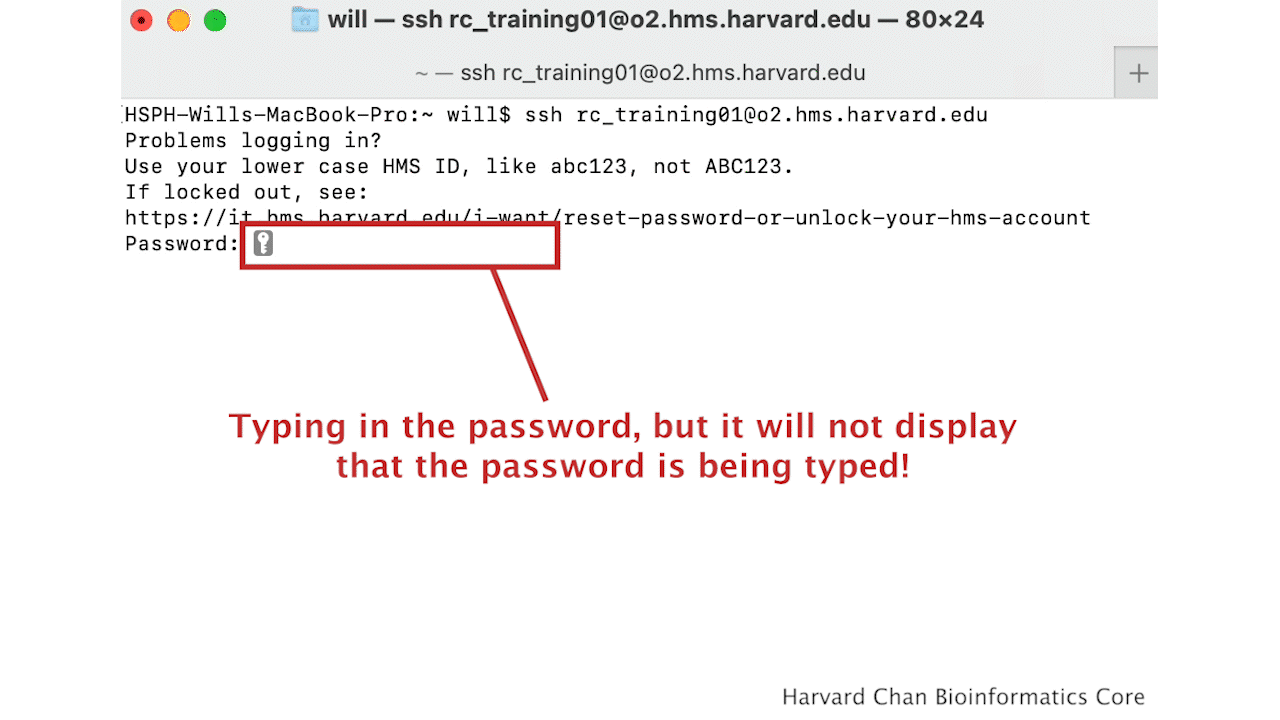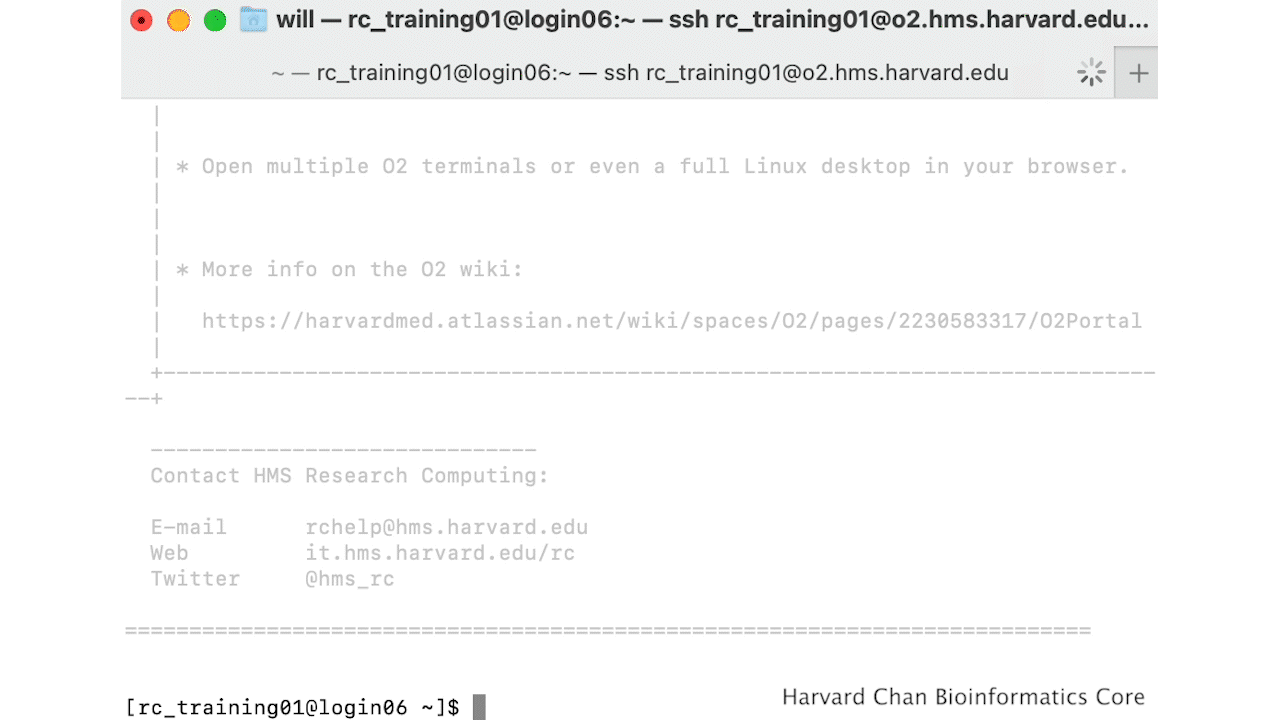
A warning might pop up the first time you try to connect to a remote machine, type “Yes” or “Y”.
Once logged in, you should see the O2 icon, some news, and the command prompt, e.g. [rc_training10@login01 ~]$.
Note 1:
sshstands for secure shell. All of the information (like your password) going between your computer and the O2 login computer is encrypted when usingssh.
About nodes:
- A “node” on a cluster is essentially a computer in the cluster of computers.
- There are dedicated login nodes and compute nodes.
- A login node’s only function is to enable users to log in to a cluster, it is not meant to be used for any actual work/computing. There are 6 login nodes on O2.
- There are several hundred compute nodes on O2 available for performing your analysis/work.
Connect to a compute node on O2
You can access compute nodes in 2 ways using a job scheduler or resource manager like Slurm.
- Directly using an interactive session (Slurm’s
sruncommand):- The
sruncommand with a few mandatory parameters will create an “interactive session” on O2. - This is essentially a way for us to do work on the compute node directly from the terminal.
- If the connectivity to the cluster is lost in the middle of a command being run that work will be lost in an interactive session.
- The
- By starting a “batch” job (Slurm’s
sbatchcommand):- The
sbatchcommand with a few mandatory parameters + a specialized shell script will result in the script being run on a compute node. - This “job” will not be accessible directly from the Terminal and will run in the background.
- Users do not need to remain connected to the cluster when such a “batch job” is running.
- The
For now let’s start an interactive session on O2 using srun.
$ srun --pty -p interactive -t 0-3:00 --mem 1G /bin/bash
In the above command the parameters we are using are requesting specific resources:
--pty- Start an interactive session-p interactive- on the “partition” called “interactive” (a partition is a group of computers dedicated to certain types of jobs, interactive, long, short, high-memory, etc.)-t 0-8:00- time needed for this work: 0 days, 8 hours, 0 minutes.--mem 1G- memory needed - 1 gibibyte (GiB)/bin/bash- You want to interact with the compute node using the bash shell
These parameters are used for
sbatchas well, but they are listed differently within the script used to submit a batch job. We will be reviewing this later in this lesson.
Make sure that your command prompt now contains the word “compute”, e.g. [rc_training10@compute-a-16-163 ~]$.
You are now working on a compute node directly in an “interactive” session!
Let’s check how many jobs we have running currently, and what resources they are using.
$ O2squeue
More about Slurm
- Slurm = Simple Linux Utility for Resource Management
- Fairly allocates access to resources (computer nodes) to users for some duration of time so they can perform work
- Provides a framework for starting, executing, and monitoring batch jobs
- Manages a queue of pending jobs; ensures that no single user or core monopolizes the cluster
Requesting resources from Slurm
Below is table with some of the arguments you can specify when requesting resources from Slurm for both srun and sbatch:
| Argument | Description / Input | Examples | Links |
|---|---|---|---|
-p |
name of compute partition | short, medium, interactive | O2 Wiki - Guidelines for choosing a partition |
-t |
how much time to allocate to job | 0-03:00, 5:00:00 | O2 Wiki - Time limits for each partition |
-c |
max cores | 4, 8 | O2 Wiki - How many cores? |
--mem |
max memory | 8G, 8000 | O2 Wiki - Memory requirements |
-o |
name of file to create with standard output | %j.out | O2 Wiki |
-e |
name of file to create with standard error | %j.err | O2 Wiki |
-J |
name of the job | Fastqc_run, rnaseq_workflow_mov10 | O2 Wiki |
--mail-type |
send an email when job starts, ends or errors out | END, ALL | O2 Wiki |
--mail-user |
send email to this address | xyz10@harvard.edu | O2 Wiki |
sbatch job submission script
An sbatch job submission script is essentially a normal shell script with the Slurm resource request specified at the top (Slurm directives) preceded by #SBATCH. Below is an example of an sbatch shell script that is requesting the following:
- the “short” partition for 2 hours
- on 4 cores (30 minutes for each core)
- using 400MiB (100MiB for each core)
DO NOT RUN
#! /bin/sh
#SBATCH -p short
#SBATCH –t 0-02:00
#SBATCH –c 4
#SBATCH --mem=400M
#SBATCH –o %j.out
#SBATCH –e %j.err
#SBATCH -J fastqc_run
#SBATCH --mail-type=ALL
#SBATCH –-mail-user=xyz10@med.harvard.edu
## Load the fastqc module
module load fastqc/0.11.5
# run fastqc (multithreaded)
fastqc -t 4 file1_1.fq file1_2.fq file2_1.fq file2_2.fq
Using software on O2
LMOD system
In the above example we want to run the FastQC tool on four files. However, before we use the fastqc command, we’ve used the command module load fastqc/0.11.5. This module load command is part of the LMOD system available on O2. It enables users to access software installed on O2 easily, and manages every software’s dependency. The LMOD system adds directory paths of software executables and their dependencies (if any) into the $PATH variable.
So, instead of using /n/app/fastqc/0.11.5/bin/fastqc as our command, we can load the module and use fastqc as the command.
Some key LMOD commands are listed below:
| LMOD command | description |
|---|---|
module spider |
List all possible modules on the cluster |
module spider modulename |
List all possible versions of that module |
module avail |
List available modules available on the cluster |
module avail string |
List available modules containing that string |
module load modulename/version |
Add the full path to the tool to $PATH (and modify other environment variables) |
module list |
List loaded modules |
module unload modulename/version |
Unload a specific module |
module purge |
Unload all loaded modules |
Note: On O2, a lot of tools used for analysis of sequencing data need to have the
gcccompiler module loaded (module load gcc/6.2.0) prior to loading the tool of interest.
Exercises
- What are the contents of the
$PATHenvironment variable? - Try running the
multiqccommand. What do you get as output? - Check if the
multiqctool is available as a module. How many versions ofmultiqcare available? - Load
multiqc/1.9. Did you have to load any additional modules? - List all the modules loaded in your environment
- Try running the
multiqccommand again. What do you get as output? - Use the
whichcommand to find out where themultiqctool is in the file system. - Check the contents of the
$PATHenvironment variable again. Any changes compared to before?
Filesystems on O2
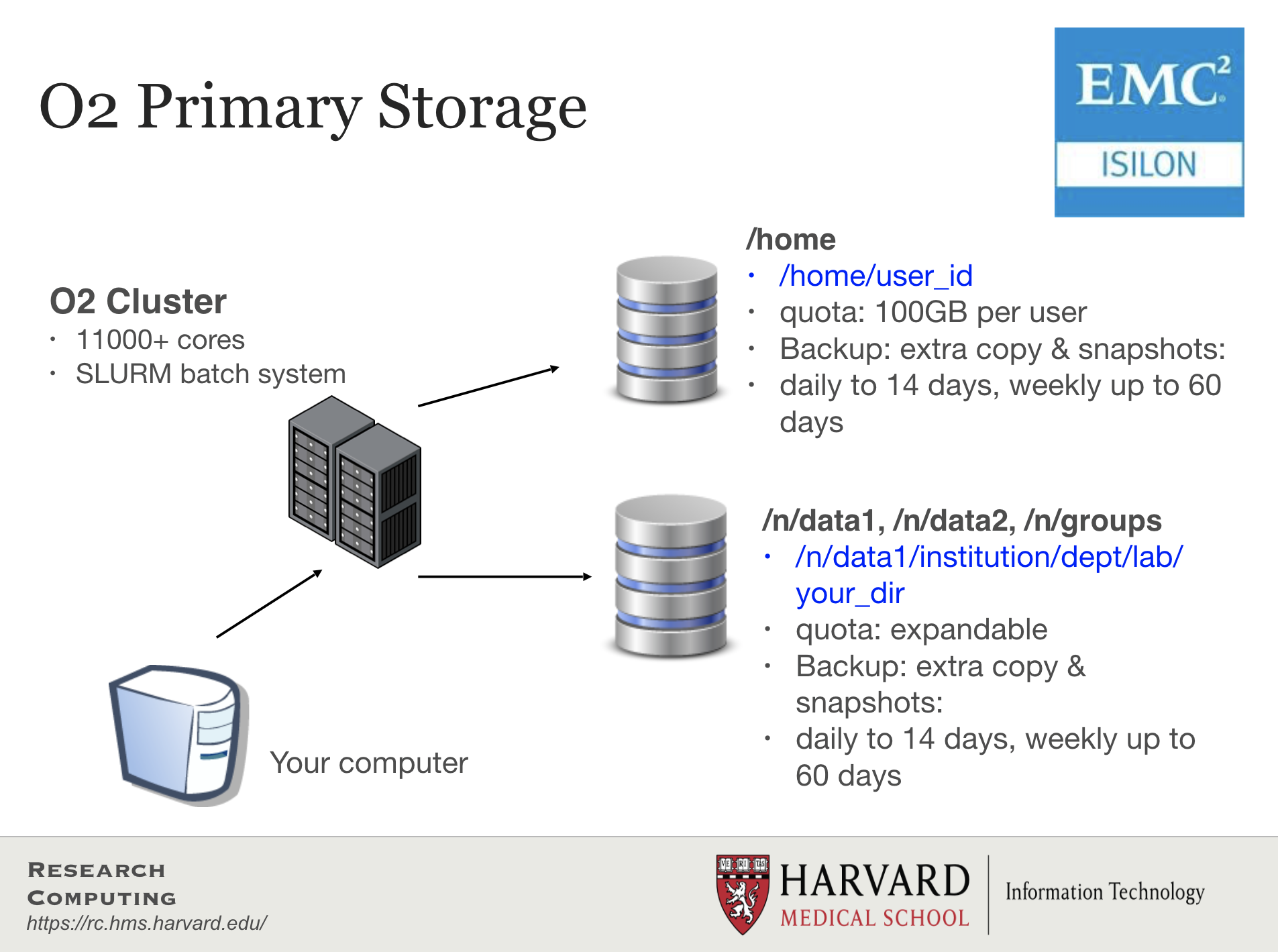
- Storage on HPC systems is organized differently than on your personal machine.
- Each node on the cluster does not have storage; instead, it is on disks bundled together externally.
- Storage filesystems can be quite complex, with large spaces dedicated to a pre-defined purpose.
- Filesystems are accessed over the internal network by all the nodes on the cluster.
- There are 3 major groups on the O2 cluster, each with their features and constraints:
/n/data1,/n/data2,/n/groups- Large datasets are stored in these parent directories (see features/constraints in the image above)./home- the home directories of all users are under this parent directory (see features/constraints in the image above)./n/scratch- scratch space for temporary storage.
Please find more information about storage on O2 by clicking here.
More about /n/scratch
- It is for data only needed temporarily during analyses.
- Each user can use up to 25 TiB and 2.5 million files/directories.
- Files not accessed for 45 days are automatically deleted.
- No backups!
- You can create your own folder using this command
/n/cluster/bin/scratch_create_directory.sh - Your scratch directory can only be created from the login node
This lesson has been developed by members of the teaching team at the Harvard Chan Bioinformatics Core (HBC). These are open access materials distributed under the terms of the Creative Commons Attribution license (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.